How to handle large datasets in pandas. We work with the FIAS database using python and 8GB of memory
There is no need to specifically represent the FIAS base:

You can download it by clicking on the link , this database is open and contains all the addresses of objects in Russia (address register). The interest in this database is caused by the fact that the files that it contains are quite voluminous. So, for example, the smallest is 2.9 GB. It is proposed to stop at it and see if pandas can cope with it if you work on a machine with only 8 GB of RAM. And if you can’t cope, what are the options in order to feed pandas this file.
Hand on heart, I have never encountered this base and this is an additional obstacle, because the format of the data presented in it is absolutely unclear.
After downloading the fias_xml.rar archive with the base, we get the file from it - AS_ADDROBJ_20190915_9b13b2a6-b3bd-4866-bd1c-7ab966fafcf0.XML. The file is in xml format.
For more convenient work in pandas, it is recommended to convert xml to csv or json.
However, all attempts to convert by third-party programs and python itself lead to a "MemoryError" error or freeze.
Hm. What if I cut the file and convert it in parts? It’s a good idea, but all “cutters” also try to read the entire file into memory and hang, python itself, which follows the path of “cutters”, does not cut it. Is 8 GB obviously not enough? Well, let's see.
You will have to use a third-party vedit program.
This program allows you to read the xml file size of 2.9 GB and work with it.
It also allows you to split it. But there is a little trick.
As you can see when reading a file, among other things, it has an opening AddressObjects tag:

So, creating parts of this large file, you must not forget to close it (tag).
That is, the beginning of each xml file will be like this:
and ending:
Now cut off the first part of the file (for the remaining parts the steps are the same).
In the vedit program:
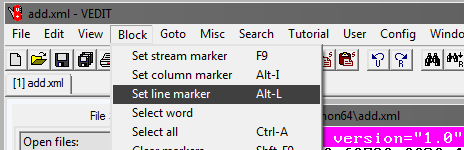
Next, select Goto and Line #. In the window that opens, write the line number, for example, 1,000,000:

Next, you need to adjust the selected block so that it captures the object in the database to the end tag:
It's okay if there is a slight overlap on the subsequent object.
Next, in the vedit program, save the selected fragment - File, Save as.
In the same way, we create the remaining parts of the file, marking the beginning of the selection block and the end in increments of 1 million lines.
As a result, you should get a 4th xml file approximately 610 MB in size.
Now you need to add tags in the newly created xml files so that they read as xml.
Open the files one by one in vedit and add at the beginning of each file:
and in the end:
Thus, now we have 4 xml parts of the split source file.
Now translate xml to csv by writing a python program.
Program code
Using the program, you need to convert all 4 files to csv.
The file size will decrease, each will be 236 MB (compare with 610 MB in xml).
In principle, now you can already work with them, through excel or notepad ++.
However, the files are still 4th instead of one, and we have not reached the goal - processing the file in pandas.
On Windows, this can be a difficult task, so we will use the console utility in python called csvkit. Installed as a python module:
* Actually this is a whole set of utilities, but one will be needed from there.
Having entered the folder with files for gluing in the console, we will perform gluing into one file. Since all the files are without headers, we will assign the standard column names when gluing: a, b, c, etc.:
The output is a finished csv file.
If you immediately upload the file to pandas
and check how much memory it will take, the result may unpleasantly surprise:

3 GB! And this despite the fact that when reading data, the first column “went” as the index column *, and so the volume would be even larger.
* By default, pandas sets its own column index.
We will carry out optimization using methods from the previous post and article :
- object in category;
- int64 in uint8;
- float64 in float32.
To do this, when reading the file, add dtypes and reading the columns in the code will look like this:
Now, by opening the pandas file, memory usage would be wise:

It remains to add to the csv file, if desired, the row-actual column names so that the data makes sense:
* You can replace the column names with this line, but then you have to change the code.
Save the first lines of the file from pandas
and see what happened in excel:

Program code for optimized opening of a csv file with a database:
In conclusion, let's look at the size of the dataset:
3.3 million rows, 28 columns.
Bottom line: with the initial csv file size of 890 MB, "optimized" for the purposes of working with pandas, it takes 1.2 GB in memory.
Thus, with a rough calculation, it can be assumed that a file of 7.69 GB in size can be opened in pandas, having previously “optimized” it.

You can download it by clicking on the link , this database is open and contains all the addresses of objects in Russia (address register). The interest in this database is caused by the fact that the files that it contains are quite voluminous. So, for example, the smallest is 2.9 GB. It is proposed to stop at it and see if pandas can cope with it if you work on a machine with only 8 GB of RAM. And if you can’t cope, what are the options in order to feed pandas this file.
Hand on heart, I have never encountered this base and this is an additional obstacle, because the format of the data presented in it is absolutely unclear.
After downloading the fias_xml.rar archive with the base, we get the file from it - AS_ADDROBJ_20190915_9b13b2a6-b3bd-4866-bd1c-7ab966fafcf0.XML. The file is in xml format.
For more convenient work in pandas, it is recommended to convert xml to csv or json.
However, all attempts to convert by third-party programs and python itself lead to a "MemoryError" error or freeze.
Hm. What if I cut the file and convert it in parts? It’s a good idea, but all “cutters” also try to read the entire file into memory and hang, python itself, which follows the path of “cutters”, does not cut it. Is 8 GB obviously not enough? Well, let's see.
Vedit program
You will have to use a third-party vedit program.
This program allows you to read the xml file size of 2.9 GB and work with it.
It also allows you to split it. But there is a little trick.
As you can see when reading a file, among other things, it has an opening AddressObjects tag:

So, creating parts of this large file, you must not forget to close it (tag).
That is, the beginning of each xml file will be like this:
<?xml version="1.0" encoding="utf-8"?><AddressObjects>
and ending:
</AddressObjects>
Now cut off the first part of the file (for the remaining parts the steps are the same).
In the vedit program:

Next, select Goto and Line #. In the window that opens, write the line number, for example, 1,000,000:

Next, you need to adjust the selected block so that it captures the object in the database to the end tag:

It's okay if there is a slight overlap on the subsequent object.
Next, in the vedit program, save the selected fragment - File, Save as.
In the same way, we create the remaining parts of the file, marking the beginning of the selection block and the end in increments of 1 million lines.
As a result, you should get a 4th xml file approximately 610 MB in size.
We finalize the xml parts
Now you need to add tags in the newly created xml files so that they read as xml.
Open the files one by one in vedit and add at the beginning of each file:
<?xml version="1.0" encoding="utf-8"?><AddressObjects>
and in the end:
</AddressObjects>
Thus, now we have 4 xml parts of the split source file.
Xml-to-csv
Now translate xml to csv by writing a python program.
Program code
here
.
# -*- coding: utf-8 -*- from __future__ import unicode_literals import codecs,os import xml.etree.ElementTree as ET import csv from datetime import datetime parser = ET.XMLParser(encoding="utf-8") tree = ET.parse("add-30-40.xml",parser=parser) root = tree.getroot() Resident_data = open('fias-30-40.csv', 'w',encoding='UTF8') csvwriter = csv.writer(Resident_data) start = datetime.now() for member in root.findall('Object'): object = [] object.append(member.attrib['AOID']) object.append(member.attrib['AOGUID']) try: object.append(member.attrib['PARENTGUID']) except: object.append(None) try: object.append(member.attrib['PREVID']) except: object.append(None) #try: # object.append(member.attrib['NEXTID']) #except: # object.append(None) object.append(member.attrib['FORMALNAME']) object.append(member.attrib['OFFNAME']) object.append(member.attrib['SHORTNAME']) object.append(member.attrib['AOLEVEL']) object.append(member.attrib['REGIONCODE']) object.append(member.attrib['AREACODE']) object.append(member.attrib['AUTOCODE']) object.append(member.attrib['CITYCODE']) object.append(member.attrib['CTARCODE']) object.append(member.attrib['PLACECODE']) object.append(member.attrib['STREETCODE']) object.append(member.attrib['EXTRCODE']) object.append(member.attrib['SEXTCODE']) try: object.append(member.attrib['PLAINCODE']) except: object.append(None) try: object.append(member.attrib['CODE']) except: object.append(None) object.append(member.attrib['CURRSTATUS']) object.append(member.attrib['ACTSTATUS']) object.append(member.attrib['LIVESTATUS']) object.append(member.attrib['CENTSTATUS']) object.append(member.attrib['OPERSTATUS']) try: object.append(member.attrib['IFNSFL']) except: object.append(None) try: object.append(member.attrib['IFNSUL']) except: object.append(None) try: object.append(member.attrib['OKATO']) except: object.append(None) try: object.append(member.attrib['OKTMO']) except: object.append(None) try: object.append(member.attrib['POSTALCODE']) except: object.append(None) #print(len(object)) csvwriter.writerow(object) Resident_data.close() print(datetime.now()- start) #0:00:21.122437
Using the program, you need to convert all 4 files to csv.
The file size will decrease, each will be 236 MB (compare with 610 MB in xml).
In principle, now you can already work with them, through excel or notepad ++.
However, the files are still 4th instead of one, and we have not reached the goal - processing the file in pandas.
Glue files into one
On Windows, this can be a difficult task, so we will use the console utility in python called csvkit. Installed as a python module:
pip install csvkit
* Actually this is a whole set of utilities, but one will be needed from there.
Having entered the folder with files for gluing in the console, we will perform gluing into one file. Since all the files are without headers, we will assign the standard column names when gluing: a, b, c, etc.:
csvstack -H fias-0-10.csv fias-10-20.csv fias-20-30.csv fias-30-40.csv > joined2.csv
The output is a finished csv file.
Let's work in pandas on optimizing memory usage
If you immediately upload the file to pandas
import pandas as pd import numpy as np gl = pd.read_csv('joined2.csv',encoding='ANSI',index_col='a') print (gl.info(memory_usage='deep')) # def mem_usage(pandas_obj): if isinstance(pandas_obj,pd.DataFrame): usage_b = pandas_obj.memory_usage(deep=True).sum() else: # , , usage_b = pandas_obj.memory_usage(deep=True) usage_mb = usage_b / 1024 ** 2 # return "{:03.2f} " .format(usage_mb)
and check how much memory it will take, the result may unpleasantly surprise:

3 GB! And this despite the fact that when reading data, the first column “went” as the index column *, and so the volume would be even larger.
* By default, pandas sets its own column index.
We will carry out optimization using methods from the previous post and article :
- object in category;
- int64 in uint8;
- float64 in float32.
To do this, when reading the file, add dtypes and reading the columns in the code will look like this:
gl = pd.read_csv('joined2.csv',encoding='ANSI',index_col='a', dtype ={ 'b':'category', 'c':'category','d':'category','e':'category', 'f':'category','g':'category', 'h':'uint8','i':'uint8','j':'uint8', 'k':'uint8','l':'uint8','m':'uint8','n':'uint16', 'o':'uint8','p':'uint8','q':'uint8','t':'uint8', 'u':'uint8','v':'uint8','w':'uint8','x':'uint8', 'r':'float32','s':'float32', 'y':'float32','z':'float32','aa':'float32','bb':'float32', 'cc':'float32' })
Now, by opening the pandas file, memory usage would be wise:

It remains to add to the csv file, if desired, the row-actual column names so that the data makes sense:
AOID,AOGUID,PARENTGUID,PREVID,FORMALNAME,OFFNAME,SHORTNAME,AOLEVEL,REGIONCODE,AREACODE,AUTOCODE,CITYCODE,CTARCODE,PLACECODE,STREETCODE,EXTRCODE,SEXTCODE,PLAINCODE,CODE,CURRSTATUS,ACTSTATUS,LIVESTATUS,CENTSTATUS,OPERSTATUS,IFNSFL,IFNSUL,OKATO,OKTMO,POSTALCODE
* You can replace the column names with this line, but then you have to change the code.
Save the first lines of the file from pandas
gl.head().to_csv('out.csv', encoding='ANSI',index_label='a')
and see what happened in excel:

Program code for optimized opening of a csv file with a database:
the code
import os import time import pandas as pd import numpy as np # : object-category, float64-float32, int64-int gl = pd.read_csv('joined2.csv',encoding='ANSI',index_col='a', dtype ={ 'b':'category', 'c':'category','d':'category','e':'category', 'f':'category','g':'category', 'h':'uint8','i':'uint8','j':'uint8', 'k':'uint8','l':'uint8','m':'uint8','n':'uint16', 'o':'uint8','p':'uint8','q':'uint8','t':'uint8', 'u':'uint8','v':'uint8','w':'uint8','x':'uint8', 'r':'float32','s':'float32', 'y':'float32','z':'float32','aa':'float32','bb':'float32', 'cc':'float32' }) pd.set_option('display.notebook_repr_html', False) pd.set_option('display.max_columns', 8) pd.set_option('display.max_rows', 10) pd.set_option('display.width', 80) #print (gl.head()) print (gl.info(memory_usage='deep')) # def mem_usage(pandas_obj): if isinstance(pandas_obj,pd.DataFrame): usage_b = pandas_obj.memory_usage(deep=True).sum() else: # , , usage_b = pandas_obj.memory_usage(deep=True) usage_mb = usage_b / 1024 ** 2 # return "{:03.2f} " .format(usage_mb)
In conclusion, let's look at the size of the dataset:
gl.shape
(3348644, 28)
3.3 million rows, 28 columns.
Bottom line: with the initial csv file size of 890 MB, "optimized" for the purposes of working with pandas, it takes 1.2 GB in memory.
Thus, with a rough calculation, it can be assumed that a file of 7.69 GB in size can be opened in pandas, having previously “optimized” it.
All Articles