
この記事では、 IDSおよびトラフィック処理システムの分野におけるソリューションの分析、攻撃の簡単な分析、IDSの動作原理の分析を提供します。 次に、ネットワークアクティビティを分析するニューラルネットワーク法に基づいて、ネットワーク内の異常を検出するモジュールを開発する試みが行われました。その目的は次のとおりです。
- コンピュータネットワークでの侵入検知。
- 輻輳および重要なネットワーク操作モードに関するデータの取得。
- ネットワークの問題とネットワーク障害の検出。
記事の目標と目的
この記事の目的は、主に問題を研究するために、応用分野でニューラルネットワークの装置を使用する試みです。
目標を実現するタスクは、通常モードからのデータネットワークの動作の逸脱を検出するためのニューラルネットワークモジュールの構築です。
記事の構造
研究と正当化:
- 侵入検知システムの種類。
- 既存のソリューションの簡単な市場調査。
- ニューラルネットワークソリューションの分野における作業の簡単な研究。
開発:
- モジュールとそのタスクの要件。
- モジュール設計。
- コンセプトの実装。
- 結論:
- コンセプトのテスト。
- 改善の機会。
- 使用済みの材料。
侵入検知システムの種類
[11]にはSOWの古典的な区分があります:
- ルーターからのトラフィックが割り当てられるネットワークレベルのシステム(ネットワークベース)。
- たとえば、ログやネットワークアクティビティを分析することにより(ホストベース)、単一のマシン上の変更を検出するホストレベルのシステム。
- 脆弱性評価システム。
この記事では、この部門をいくらか拡大したいと思います。
実際、そのようなシステムの目標は質問に答えることです。問題はありますか?
決定は、受信したデータに基づいて行われます。
つまり システムタスクは以下で構成されます:
- データを受信しています。
- 取得したデータの解釈。
- 結果の表現。
したがって、すべてのシステムは、次の機能の値に従って配置できます。
- 収集されたデータのタイプ。
- データを取得する方法。
- データ解釈方法
- 結果を提示する方法。
非体系的な特性、結果に対する反応のタイプを考えます:
- 情報提供。
- アクティブ
最初のケースでは、利害関係者に通知されます。
2番目-アクティブなアクション、たとえば、攻撃者のアドレス範囲のブロック。
これに基づいて、これらのシステムは通常、人為的にIDSとIPSに分割されます。
私はシステムのパーティションを想定しているため、この特性は非体系的です
「インテリジェンス」と「パワー」ユニットに。 また、IPSにIDSを含めることができます。
さらに、この質問には戻りません。
データ型クラス
古典的な分割に続いて、2つのクラスを導入し、3つ目のクラスを追加して、クラスを「構成スペースの座標」として提示できるようにします(後ですべての順列をすばやく取得するためだけです)。
- ホストについて収集されたデータ。
- ネットワーク全体について収集されたデータ。
- ハイブリッドシステム。
ホストについて収集されるデータのタイプは、1つのホストのみに関係するデータであり、部分的にはホストと対話するデータに関係します。
このようなデータの分析により、「このホストに攻撃はありますか?」という質問に答えることができます。
原則として、このデータをノードで直接収集する方が便利ですが、これは必須ではありません。
たとえば、ネットワークスキャナは、外部から特定のホストで開いているポートのリストを取得できますが、そこではコードを実行できません。
このクラスには、次のタイプのデータが含まれます(それぞれに収集された特定のインジケーターが含まれます)。
- ノードのネットワークアクティビティ。
- ノードのネットワーク設定。
- ファイルデータ(リストとチェックサム、メタデータ、ファイルアクションなど)。
- プロセスデータ。
同時に、ノードは、サービスを提供するサーバーとして使用することを意図していないワークステーションとサーバーの両方にすることができます。
別のケースを強調したいので、ホストをタイプに分けます。攻撃方法(ニューラルネットワークのトレーニングを含む)を学習し、攻撃ノードを識別するために、ホストを特別に脆弱にすることができます。
このノードとの相互作用は攻撃の試みであると想定することができます。
ネットワークについて収集されたデータは、ネットワーク接続の全体像です。
原則として、完全なネットワークデータは収集されません。 リソースを集中的に使用するため、侵入者はネットワーク内にいられないか、外部と通信する必要があると考えられます(もちろん、物理的に隔離されたネットワークでも攻撃は可能です。 )
この場合、IDSはルーターを通過するトラフィックを分析します。ルーターには、トラフィックがIDSにリダイレクトされるSPANポートがあります。
原則として、IDSを実行しているホストからデータを収集することを妨げるものは何もありません。さらに有用であり、追加の制御をもたらします。
ノードでネットワークトラフィックを収集することもできます。 ただし、これにより、ホストネットワークアダプターはすべてのトラフィックをキャプチャするモードで動作するようになりますが、これは通常の運用では通常予期されないことであり、これは明らかに冗長です(ストレッチで有用な場合がある唯一のオプションは分散フロー分析です)。
データメソッドクラス
データ取得方法のクラス:
- パッシブ。 システムはネットワークパフォーマンスに直接影響しません。 トラフィック分析を行うだけです。
- アクティブ システムは、たとえば、応答トラフィックでなじみのあるシグネチャを見つけるために、ネットワークに積極的に影響を与える「戦闘中の偵察」を実行しようとします。
- 混合。 上記の両方のクラスが使用されます。
パッシブ検出
パッシブ検出では、システムは状況を監視するだけです。 ほとんどのIDSはこのクラスのメソッドを使用します。 ホストレベルのシステムも通常、このクラスのメソッドを使用します。 たとえば、ユーザーの下からシステムファイルを削除して削除されたことを確認するのではなく、単にこのファイルに対する権限がデータベース内のテンプレートに準拠しているかどうかを評価し、準拠していない場合は警告を発行します。
アクティブな脆弱性スキャン
このクラスのメソッドでは、既知および未知の両方のアクション(ファジーシステム)によってエラーが引き起こされます。
その後、データベースはこれらのアクションに対する反応を分析します。 このクラスのメソッドは、脆弱性スキャナーに一般的です。
結果を解釈するには、両方のクラスのメソッドが適用可能です。
- データベースに基づく回答の分析(たとえば、一般的なSQLインジェクションパターンへの一般的な回答)、または行動分析(その後の目標の動作方法、および次のクエリへの回答方法)にはさまざまなものがあります。
たとえば、誤って構成されたIPパケットを送信すると、脆弱なサーバーがクラッシュし、その後応答しなくなります。 - 異常なアクティビティを検出するオプションがあります。
たとえば、0xDEADBEEFが満たされた777バイトのICMPパケットを送信した後、ネットワークアクティビティレベルが急激に増加してから低下した場合-これは異常です(通常の状況-ネットワークアクティビティのレベルは変化しません)。
利点は明らかです:
- 積極的な侵入検知。
攻撃は発生しませんが、ネットワークは攻撃に対して脆弱なままです。
このクラスのメソッドは、潜在的な脆弱性を識別します。 - ネットワークスキャンは攻撃者に似ており、脆弱性を検出する可能性が高くなります。
短所:
- 追加のネットワーク負荷。
- 一部のサービスのDoSなど、スキャンプロセス中に成功した攻撃を実装する機能。
- 原則として、それは攻撃のベースに依存し、攻撃は時代遅れになりつつあります。
- 明らかなマイナスは、セキュリティの幻想です。 「スキャナーが何も検出しなかった場合、すべては問題ありません」と感じるかもしれません。
同時に、標的型攻撃のリスクは依然として残っています。 このリスクは、おそらく程度は低いですが、他のクラスのメソッドで発生します。
データ解釈メソッドクラス
次のクラスを区別することができます。各クラスにはいくつかのメソッドが含まれる場合があります。
- 既知の違反の検出方法。
- 異常を検出する方法。
- 上記の両方を含む混合メソッド。
既知の違反の検出
既知の攻撃の兆候を探すことになります。
利点:
- メソッドはほとんど誤検知の傾向がありません。
- 原則として、サンプルとの比較として実装されたメソッドは、非常に迅速に実行され、リソースの大きな支出を必要としません。
これらの方法の欠点は、システムにとって未知の攻撃を検出できないことです。
クラシックバージョンでは、パケット署名とデータベース内の署名を比較することが実装に含まれます。 比較は、パターンを使用するか、正規表現を使用して正確にすることができます。
最もよく知られているIDSは、このクラスのメソッドを使用します。
ここにファジー比較方法を含めることもできます。
- 訓練されたパーセプトロン。
- ファジーロジックを使用した比較方法。
異常検出
一番下の行は、通常のネットワークアクティビティのパターンを記録し、このパターンからの逸脱に対応することです。
ネットワーク上の「通常の」パケットの署名のデータベース形式での可能な実装
アナライザーがいくつかのまれなアクションまたはアクティビティを検索するときの統計的偏差検出システム 。 システム内のイベントは統計的に検査され、症状が異常と思われるイベントを見つけます。
以下の検出器は、異常の検出を試みます。
結果表示方法
既存のソリューションの分析が示しているように、通常2種類の方法が使用されます。
- 2クラス。 システムは、「問題はありますか?」という質問に答え、「はい/いいえ」という答えで答えます。
この方法は、学術論文や研究では一般的ですが、実際のSOWのモジュールやセンサーの作成にも適用できます。 - マルチクラス。 システムは、「現在問題は何ですか?」という質問に答えます。 問題がなければ、何もありません。
結果は一定の自信を持って提示することもできます。 また、ニューラルネットワークは結果を「はい」または「いいえ」ではなく、結果の確率のセットとして表します。
既存のアプローチでは、確率は1または0に概算され(たとえば、ネットワークの出力で起こりうるすべての攻撃から、最初の確率が最も高い確率で選択されます)、他のどこにも考慮されません。
IDSタイプ
この段階では、パラメータ「結果表示方法」は重要ではありません(検出器を分離するために導入されました)。
from itertools import product data_type_class = ('host_data', 'net_data', 'hybrid_data') analyzer_type_class = ('passive_analyzer', 'active_analyzer', 'mixed_analyzer') detector_type_class = ('known_issues_detector', 'anomaly_detector', 'mixed_detector') liter = list(product(data_type_class, analyzer_type_class, detector_type_class)) print(len(liter)) for n, i in enumerate(liter): print('Type {}:\ndata_type_class = {}, analyzer_type_class = {}, detector_type_class = {}.'.format(n, i[0], i[1], i[2])) print('-' * 10)
次に、それらをより少ないタイプに減らします。
- タイプ:0、1、2。 署名検索、異常な動作の検出、またはハイブリッドに基づく ノードレベルのIDS 。 同様のIDSが多数あります。 アンチウイルスもここに含まれている場合があります。
- タイプ:3、4、5。署名検索、異常な動作の検出、またはハイブリッドに基づくノードレベルの脆弱性スキャナー 。 オープンな脆弱性のスキャナー。 この機能はアンチウイルスに組み込まれています。 ストレッチを使用して、グループの代表として、 AVZを強調表示することができます。 さらに、このタイプを個別に解析しません。
- タイプ: 6、7、8 。 ハイブリッドホストレベルIDS 。攻撃を受動的に追跡し、アクティブスキャンを可能にします。
- タイプ: 9、10、11。IDSネットワークレベル 、署名の検索に基づいて、異常な動作を検出、またはハイブリッド。
- タイプ:12、13、14。シグネチャ検索、異常反応の検出、またはハイブリッドに基づくネットワークレベルの脆弱性スキャナー 。
- タイプ:15、16、17。異常な動作を検出するハイブリッドネットワークレベルID 、またはハイブリッド。 パッシブモードでのスキャンと検出の両方の問題を許可する必要があります。 これらは完全なネットワークレベルのソリューションです。 通常、これらはこのフォームには存在せず、次のタイプとマージされます。
- タイプ26(タイプ18から25は通常、個々に意味をなさないため、このタイプに縮小されます)。 エージェントに基づくハイブリッドシステム 。各エージェントは上記のタイプのいずれかを持つことができます。
- IDSではないが、補助システムとして行われるため、分類に分類されない別個のアイテムはhoneypotです。
以下は、上記のグループに分類される既存のソリューションの例です。
既存のソリューション
既存のシステム
鼻水
Snortは、従来のネットワークレベルのIDSであり、トラフィックを分析してルールベース(実際には署名ベース)に一致します。 つまり、このシステムは既知の違反を探しています。 Snortがそのモジュールを簡単に実装することは可能ですが、これはいずれかの作業で行われました。 ロシアのものを含む 、Snort に基づいた多くの有名な商用ソリューションがあります 。
Snort IDSに基づいて構築された署名データベースの操作に加えて、ヒューリスティック、ニューラルネットワーク、および同様の検出モジュールが含まれることがあります。 少なくとも、Snort用の機能する統計的な異常検出機能があります。
すりかた
Suricataは、Snortと同様、ネットワークレベルのシステムです。
このシステムにはいくつかの機能があります。
- 署名ベースはSnortと互換性があります。
- ネットワーク/トランスポート層だけでなく、アプリケーションプロトコルのレベルでも機能します。
- 可能性の範囲を広げるインタープリター言語であるLuaでルールを実装することが可能です。
- 一般に、個々のパケット/接続だけでなく、2つのホスト間のトラフィックを分析できます。 これにより、たとえば、パスワードを推測する試みを検出できます。
- 各IPアドレスに「レピュテーションレベル」を割り当てることができるIPレピュテーションサブシステムがあります。
つまり、このシステムは、以前の違反と同様に既知の違反を検出しますが、適応性と学習能力が高くなります(ホストの評価レベルはシステムの運用中に変化し、意思決定に影響を与える可能性があります)。
ブロ
ネットワークレベルのIDSを作成するためのプラットフォーム。 これは、既知の違反の検出に重点を置いたハイブリッドシステムです。 トランスポート、ネットワーク、およびアプリケーションレベルで機能します。 独自のスクリプト言語をサポートします。
たとえば、異なるポートのサービスへの複数の接続などの異常を検出することができます-通常のホストでは一般的ではない動作が検出されます。
これは、最初に、送信されたデータの正常性のチェックに基づいて実現されます(たとえば、 すべてのフラグが設定されたTCPパケットは、正しいにもかかわらず、おそらくここで何かが間違っています)。
次に、ネットワークが正常に機能する方法を説明するポリシーに基づいています。
Broは攻撃を検出するだけでなく、ネットワークの問題の診断にも役立ちます(機能の主張)。
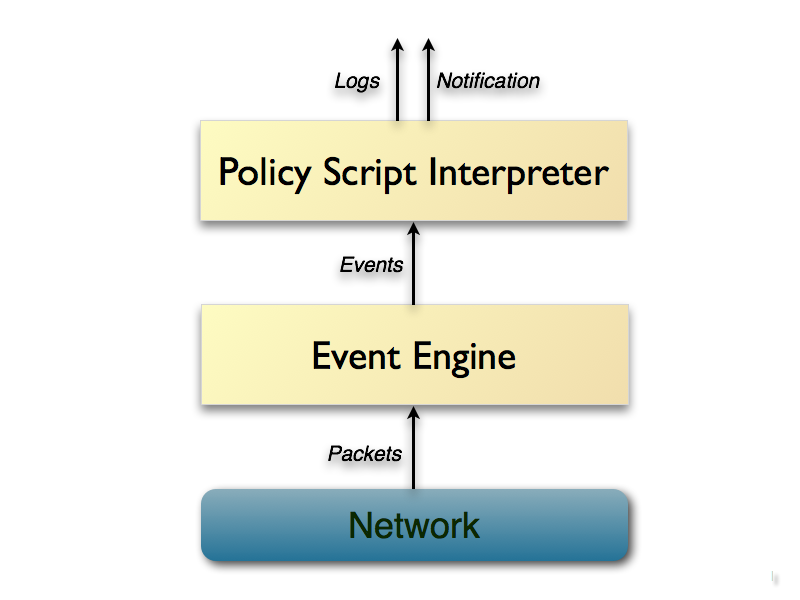
技術的には、Broは非常に興味深い実装です。標識によってトラフィックを直接分析するのではなく、「イベントマシン」でパケットを実行し、パケットストリームを一連の高レベルイベントに変換します。
このマシンは、中立のポリシー(イベントは単に何かが発生したことを通知しますが、なぜ、イベントエンジンは何も言わない、t .k。これはポリシーインタープリターのタスクです)。
たとえば、HTTP要求は、適切なパラメーターを使用してhttp_request
イベントに変換され、分析レベルに渡されます。
ポリシーインタープリターは、イベントハンドラーがインストールされているスクリプトを実行します。 これらのハンドラーでは、トラフィックの統計パラメーターを計算できます。 同時に、ハンドラーは単一のパッケージに応答するだけでなく、コンテキストを維持できます。
つまり、時間のダイナミクス、フローの「履歴」が考慮されます。
Broコアの動作の簡単な図:
TripWire 、 OSSEC 、 Samhain
古典的な用語では、これらはホストレベルのシステムの代表です。
既知の違反を検出する方法と異常なアクティビティを検索する方法を組み合わせます。
異常検索メカニズムは、インストール中にシステムがシステムファイルのハッシュとそれらに関するメタ情報をデータベースに保存するという事実に基づいています。 オペレーティングシステムパッケージを更新するとき、ハッシュが再カウントされます。
監視対象ファイルの制御されていない変更の場合、しばらくするとシステムから通知されます(通常、FSイベントに応答するシステムは可能ですが、スキャンはスケジューラによって開始されます)。
ファイルの監視に加えて、これらのシステムはプロセス、接続を監視し、システムログを分析できます。
また、定期的に更新される既知の違反のデータベースを使用できます。
一部には、多数のノードで同時にデータを分析できる集中化されたインターフェイスがあります。
良い点はSamhainのWebサイトで説明されています。
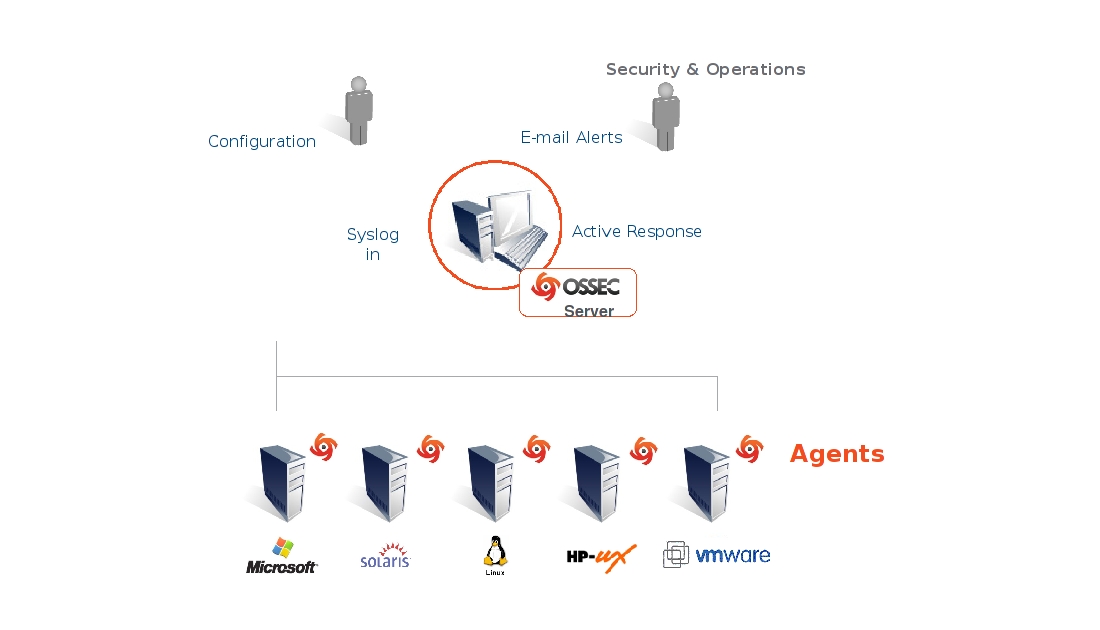
Samhainホストベースの侵入検知システム(HIDS)は、ファイルの整合性チェックとログファイルの監視/分析、ルートキットの検出、ポートの監視、不正なSUID実行可能ファイルの検出、隠しプロセスを提供します。
Samhainは、潜在的に異なるオペレーティングシステムで複数のホストを監視するように設計されており、単一のホストでスタンドアロンアプリケーションとして使用することもできますが、集中ログとメンテナンスを提供します。
OSSEC展開の例:
プレリュードSIEM 、 OSSIM
これらは、 SIEMとして位置付けられるハイブリッドシステムです。 Preludeは、センサーネットワークとアナライザーを組み合わせたものです。 攻撃者は1つのIDSをバイパスできるため、このシステムはセキュリティレベルを向上させると述べられていますが、多数の保護メカニズムをバイパスする複雑さが指数関数的に増加します。
システムはうまく拡張できます。開発者によると、センサーネットワークは大陸または世界をカバーできます。
システムは、AuditD、Nepenthes、NuFW、OSSEC、Pam、Samhain、Sancp、Snort、Suricata、Kismetなどを含む、データ形式(IDMEF)による多くの既存のIDSと互換性があります。
OSSIMについてもほぼ同じことが言えます。
脆弱性スキャナー
これらは、ホストまたはネットワーク上の脆弱性を積極的に検索するシステムです。
データベース内の既知の問題のみを検索する最も単純な方法では、より深刻なシステムでは、既知の違反と異常なアクティビティを検出する両方の方法を組み合わせることができます。
脆弱性スキャナーはかなり前に登場し、非常に多くのことに成功しました。
市場に出回っている有名な製品をいくつか紹介します。
- Nmapは、補助ツールとしてより使用される無料のネットワークスキャナーです。OSの識別、開いているポートとそれらを開いたサービスのリストの取得などができます。同時に、Netmap Scripting Engineが含まれており、自動化されたものを構築できますソリューション(たとえば、スキャンを必要なポートでのbrutefoserの起動と組み合わせる)。
- XSpiderは、データベース、ヒューリスティック、異常動作検索などを備えた古典的な脆弱性スキャナーです。
- Metasploit-エクスプロイトキット、スキャナー、攻撃されたノードで起動される既製のロードなどを含むフレームワーク...既知の脆弱性にさらされているノードまたはノードのグループを調査できます。
- OpenVASは、スキャナーと脆弱性管理ソリューションを組み合わせたフレームワークです。 50,000を超える脆弱性を持つデータベースが含まれ、指定されたターゲットを識別し、対応するエクスプロイトの使用に対する反応をチェックします。
上記のXSpiderの例におけるこれらのシステムの機能:
- 非標準構成のサーバーの脆弱性を識別するためのランダムポート上のサービスの識別。
- サービスのタイプと名前(HTTP、FTP、SMTP、POP3、DNS、SSHなど)を決定するための発見的方法で、実サーバー名とチェックの正しい操作を決定します。
- RPCサービス(Windowsおよび* nix)の処理(コンピューターの詳細な構成の決定を含む)。
- パスワード保護の脆弱性の確認:認証を必要とするほとんどすべてのサービスでのパスワード選択の最適化。
- スクリプトの脆弱性の検出を含む、Webサイトコンテンツの詳細な分析:SQLi、XSS、任意のプログラムの起動など。
- HTTPサーバーの構造を分析して、構成の弱点を見つけます。
- Windowsでの高度なサイトスキャン。
- 非標準のDoS攻撃のチェックを実施します。
- 自動化機能。
ハニーポット
システムは、攻撃を誘引するために特別に脆弱になりました。 攻撃者の行動を研究するために使用できます。
展開例:
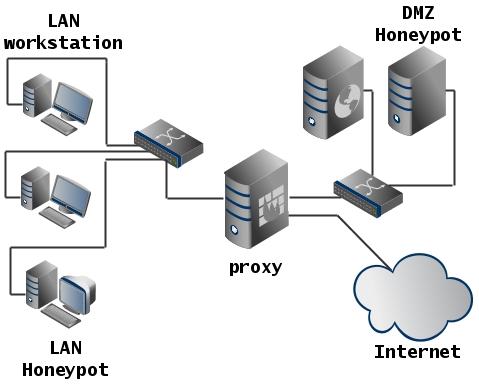
一般に、多くの既製のハニーポットソリューションがあります。
これには、ハニーポットの「アクティブ」バージョンと見なすことができる、攻撃を遅くする(攻撃者のリソース要件を増やし、応答に時間を与える) tarpitなどのトラップも含まれる場合があります。
問題を解決するためのアプローチのバリエーションとニューラルネットワークソリューション
[14]では、広く使用されているKDDデータベースへの参照の分析が行われました 。
- サポートベクターマシン-24。
- 決定的な木-19。
- 遺伝的アルゴリズム-16。
- 主成分の方法-13。
- 粒子群法-9。
- k最近傍の検索-9。
- k値によるクラスタリング-9。
- 単純ベイズ分類器-9。
- ニューラルネットワーク、多層パーセプトロン-8。
- 遺伝的プログラミング-6。
- ラフセット -6。
- ベイジアンネットワーク-5。
- ランダムツリーのフォレスト-5。
- 人工免疫システム-5。
- ファジールールマイニングの使用-4。
- ニューラルネットワーク(自己組織化マップ)-4。
ご覧のとおり、2015年から2016年の作業中のニューラルネットワークはそれほど広く表現されていません。また、これらは通常、直接配信ネットワークです。
実際には、上記のソリューションを含むソリューションは、主に次の分析テクノロジーに依存しています。
- 既知の違反を検出する方法:
- 署名分析。 データ署名、または動作署名と更新されたデータベースの署名の比較。 署名は、パターンまたは正規表現で表すこともできます。
- ルールに基づくエキスパートシステム[1]。
- 異常検出方法:
- しきい値検出器(たとえば、サーバーのCPU負荷の安定した超過に反応する)[1]。
- 統計システム(たとえば、 ベイズ分類器 、または訓練された分類器のシステム )。
- 行動分析(ファジールールに起因する可能性があります)。
- 人工免疫システムモデルの使用 。
私が触れていないいくつかのソリューションについては、 こちらもご覧ください 。
これらのソリューションには独自の問題があります。
- シグニチャ検索は未知の攻撃に応答せず、攻撃のシグニチャの非常に大きな変更では十分ではないため、検出器はそれを検出しません。
- ルールに基づくエキスパートシステムおよび署名検索の場合、データベースの関連性を維持する必要があります。
- ルールベースのシステムの場合、攻撃中のアクションのシーケンスのわずかな変動は、適切な検出メカニズムによって攻撃が検出されない程度まで、アクティビティとルールの比較プロセスに影響を与える可能性があります。 このようなシステムで注意散漫のレベルを上げると、この問題の部分的な解決策が得られますが、これにより誤検出の数が大幅に増えます[10]。
- ルールベースのシステムには、ルールの構造に十分な柔軟性がないことがよくあります。
- 統計システムは、イベントのシーケンスに敏感ではありません(既存のすべてのシステムに当てはまるわけではありません)。
- それらおよびしきい値検出器が、攻撃検出システムによって監視される特性のしきい値を設定することは困難です。
- 時間が経つにつれて、攻撃者が正常と見なされるように、統計システムが侵入者によって再訓練される可能性があります。
ニューラルネットワークはまず第一に、異常の統計的検出器またはそれらへの追加の代替として考えられるべきですが、ある程度までは、シグネチャ検索や他の方法を置き換えることができます。
さらに、近い将来にハイブリッドシステムが使用される可能性があります。
ニューラルネットワークには、次の利点があります。
- 不完全な入力データまたはノイズの多い信号を分析する機能。
- 知識を形式化する必要はありません(トレーニングに置き換えられます)。
- フォールトトレランス:一部のネットワーク要素の障害や通信の破壊により、ネットワークが常に完全に機能しなくなるわけではありません。
- 作業の単純な並列化の可能性。
- ニューラルネットワークでは、オペレーターの介入が少なくて済みます。
- 未知の攻撃を検出する可能性があります。
- ネットワークは自動的に、またその過程で学習することができます。
- 複雑さを大幅に増加させることなく、多次元データを処理する機能。
欠点もあります:
- ほとんどのアプローチはヒューリスティックであり、多くの場合、明確なソリューションにつながりません。
- 神経症に基づいてオブジェクトのモデルを構築するには、ネットワークの予備トレーニングが必要であり、計算と時間のコストが必要です。
- ネットワークをトレーニングするには、トレーニングとテストサンプルを準備する必要がありますが、これは必ずしも簡単ではありません。
- 場合によってはネットワークトレーニングはデッドロックにつながります。ネットワークは再トレーニングの対象になるか、収束しない場合があります。
- ネットワークの動作は常に明確に予測できるとは限らないため、誤検知や見逃しのリスクが生じます。
- ネットワークがこの決定をした理由を説明するのは困難です(言語化の問題)。
- したがって、結果の再現性と明確性を保証することは不可能です。
ニューラルネットワークに基づく既存のソリューションと提案
文献に記載されているアプローチを要約します。
- Mustafayev [2]、Zhigulin and Podvorchan [6]、Halenar [7]の作品では、以前に攻撃に基づいてトレーニングされた多層パーセプトロン(KDDなど)を使用することが提案されています。
- Haibo Zhang [3]と共著者は、ニューラルネットワークとウェーブレット変換の使用を提案しています。
- Min-Joo Kang [4]と共著者は、車両のオンボードCANネットワークの問題を検出するために詳細なトレーニングを使用しています。
- Talalayev et al。[8]は、再循環ニューラルネットワークまたは主成分法を使用して特徴空間を圧縮することを提案し、その後、2層パーセプトロンとKohonenマップの両方の使用を研究しています。
- Balakhontsev et al。[10]は、3層のパーセプトロンを使用しています。
- KornevとPylkin [13]は既存のアプローチを研究し、異なる数の層を持つパーセプトロン、通常状態を検出するための単層分類器、およびKohonenマップとパーセプトロンから成るハイブリッドネットワークを使用する可能性を指摘します。
そして実際に使用される:
- DeepInstinctはディープラーニングソリューションを提供しますが、彼らが使用するテクノロジーの詳細と比較は見つかりませんでした。 サイトにあるものから判断すると、教師とのトレーニングを使用します。
- 非公式データによると、ニューラルネットワーク検出器の開発は、ロシアの数社を含む企業によって積極的に行われています。
既存のソリューションに関する結論
SAPの管理および監視ツールは、 統合ソリューションに向けて進化しています 。
最新のシステムは、全体として、侵入検知という狭いタスクを実行するだけでなく、異常検出方法と既知の違反検出方法の両方を実現しながら、ネットワーク障害の診断にも役立ちます。
このような統合システムの構成には、ネットワーク上に分散されたさまざまなセンサーが含まれ、パッシブまたはアクティブのいずれかになります。 IDSのインスタンス全体である可能性があるセンサーデータを使用して、(Prelude SIEMの用語では)中央相関器がネットワークの全体的なステータスを分析します。
同時に、処理する必要があるデータの量とそのディメンションが増加します。
文献では、主に、パーセプトロンまたはコホーネンマップに基づくニューラルネットワーク手法が検討されています。 競合するニューラルネットワークまたは畳み込みネットワークに関する最新の研究に基づいて、開発という形で満たされていないニッチがあります。 これらの方法は、複雑な分析が必要な関連分野ではうまく機能していますが。
市場のニーズと既存のソリューションの形での供給との間には明らかにギャップがあります。
したがって、この方向で研究を行うことは理にかなっています。
モジュール設計
考えられる攻撃
, , , [16].
- , , IDS ( , ARP , Ethernet, PPP , ).
, 90- — 2000-, .
Ping of the Death , IP Spoofing , SYN flood ARP cache poison , - DNS .
(, SQL injection PHP injection ), , XSS CSRF , , .
. ( , ).
Active Directory , "-" SMB .
HTTPS HSTS , SSL stripping .
, (, , HeartBleed ), .
, .
DoS , , .
(, SYN Flood) , , (, HTTP ) DDoS.
.
- , . , , DoS: , Windows- , SSH . , .
, "" (, , ftpd, sendmail ), . , : , pdf , ...
IDS : , , . , , , .
. , .
, : , , .
, , , .
- , (, : , — , ). ( , ). .
:
- .
- , , .
- , , .
, .
" ", ICMP ping. .
— TCP SYN .
, IDS . , .
, , :
- , . : .
- . , ( ).
.
:
- .
- .
- /.
- (, ).
- .
- ( , ).
- (, ).
, , , , ( : , , ).
. , , , CPU.
, .. . , , .
, - , , , , , ( "" ). , (, , ), IDS.
:
- : DoS/DDoS.
- .
- . , .
.
スキャン
:
- : n m . , , .
- : n m p .
IDS: , .
, .
, .
, , :
- CPU ().
- IO ( ).
- ( ).
- ( ).
.
[12] :
- TCP , , IP .
- ICMP , IP ,
ICMP_ID
. - IP , UDP .
[6] , NSL-KDD .
-
duration
— . -
protocol_type
— : TCP, UDP .. -
service
— : HTTP, FTP, TELNET .. -
flag
— : . -
scr_bytes
— . -
dst_bytes
— . -
land
— 1 / /. -
wrong_fragments
— "" . -
urgent
— (urgent) . -
hot
— "" . -
num_failed_logins
— . -
logged_in
— 1 — , 0 . -
num_compromised
— . -
root_shell
— 1 — , 0 — . -
su_attempted
— 1 — "su root", 0 — . . -
num_root
— "root". -
num_file_creations
— . -
num_shells
— " ". -
num_access_files
— . -
num_outbound_cmds
— ftp . -
is_host_login
— 1 — "host" . -
is_quest_login
— 1 — "". -
count
— 2 . -
srv_count
— 2 .. -
serror_rate
— syn . -
srv_serror_rate
— syn . -
rerror_rate
— rej . -
srv_rerror_rate
— rej . -
same_srv_rate
— . -
diff_srv_rate
— . -
srv_diff_hast_rate
— . -
dst_host_count
— , . -
dst_host_srv_count
— , . -
dst_host_same_srv_rate
— , . -
dst_host_diff_srv_rate
— , . -
dst_host_same_src_port_rate
— . -
dst_host_srv_diff_host_rate
— . -
dst_host_serror_rate
— c syn -. -
dst_host_srv_serror_rate
— c syn . -
dst_host_rerror_rate
— c rej -. -
dst_host_srv_rerror_rate
— c rej .
-
duration
— (). -
protocol_type
— (TCP, UDP, .). -
service
— (HTTP, TELNET .). -
flag
— c . -
src_bytes
— . -
dst_bytes
— . -
land
— 1 ; 0 . -
wrong_fragment
— "" . -
urgent
— URG.
, .
, .
- ID protocol — , .
- Source port — TCP UDP.
- Destination port — TCP UDP.
- Source Address — IP .
- Destination Address — IP .
- ICMP type — ICMP .
- Length of data transferred — .
- FLAGS — .
- TCP window size — TCP .
, , , .
, NSL-KDD , . , . , , .
, [6] .
-
duration
— . -
protocol_type
— (TCP, UDP, .). -
service
— (HTTP, TELNET .). -
flag
— c . -
src_bytes
— . -
dst_bytes
— . -
land
— 1 ; 0 . -
wrong_fragment
— "" . -
urgent
— URG. -
count
— 2 .. -
srv_count
— 2 .. -
serror_rate
— syn . -
diff_srv_rate
— . -
srv_diff_host_rate
— . -
dst_host_srv_count
— , .
IDS
IDS :
:
- .
- .
- .
:
- .
- .
- :
- .
- .
IDS, , . Prelude SIEM OSSIM, . , .
- – .
- – .
- – .
- – , .
.
. , , , .
, , .
, .
, , . , , , (, ), .
:
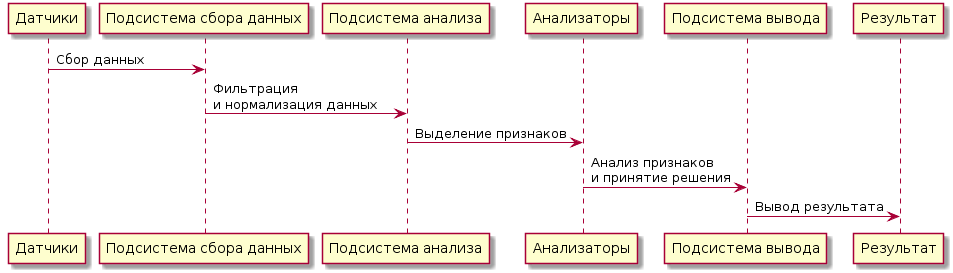
-> " ": " " -> " ": \n " " -> "": "" -> " ": \n " " -> :
, : . . , IDS .
, , . , , , ( NSL-KDD). .
, ( , ), .
, , , .
, .
, (), , , , .
, :
, , , " ".
: , , .
- , , : .
( , , ).
, , , .
, ( NSL-KDD):

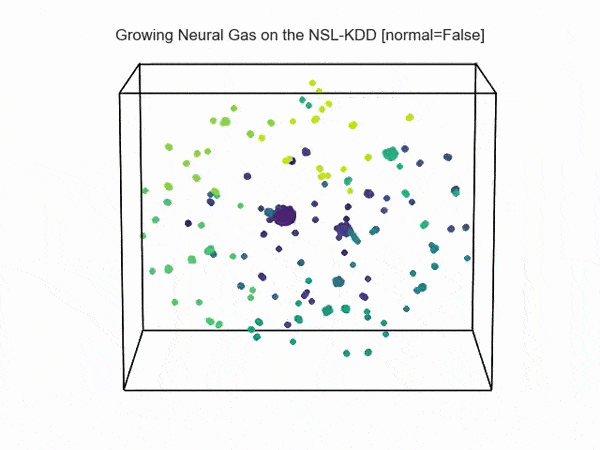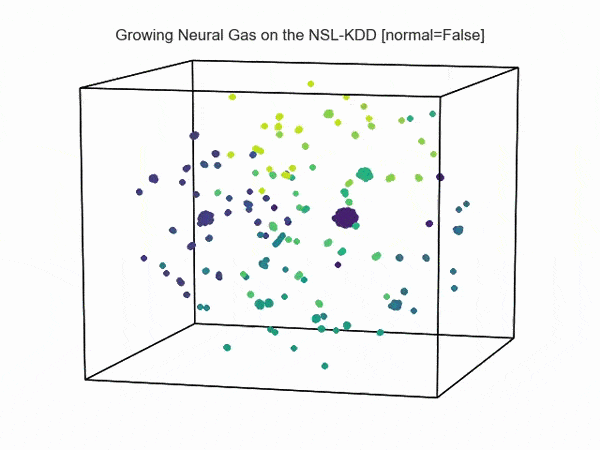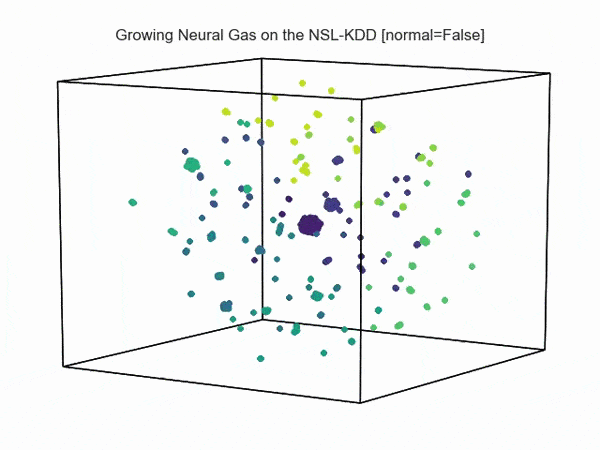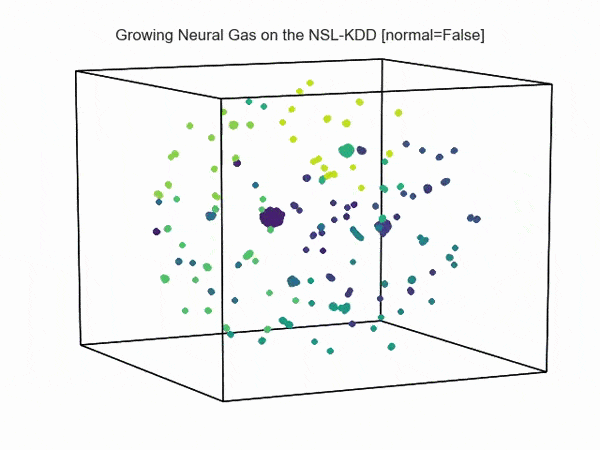
.., :
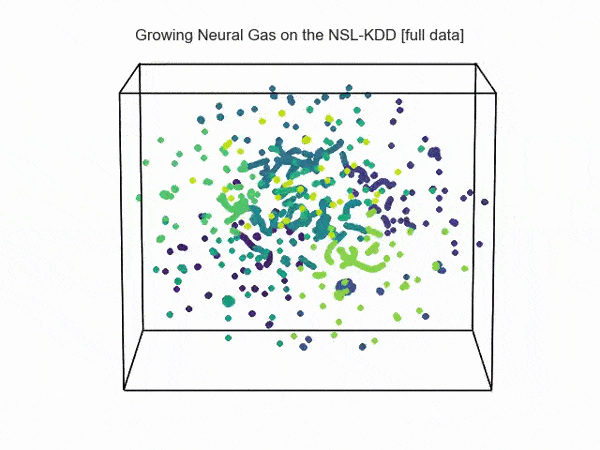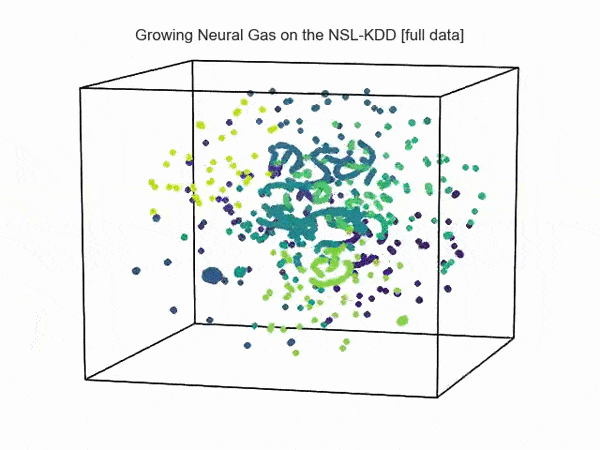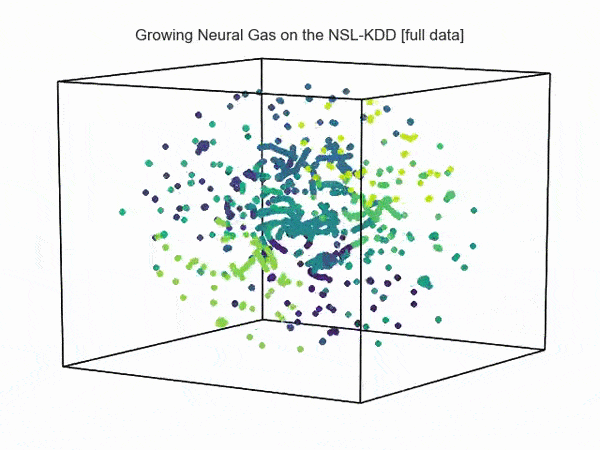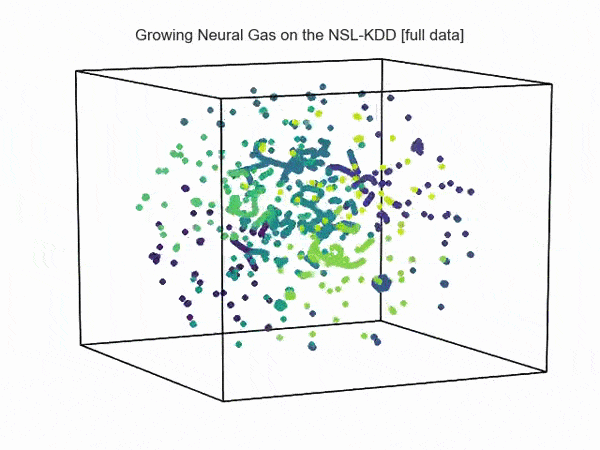
import hypertools as hyp from collections import OrderedDict import csv def read_ids_data(data_file, is_normal=True, labels_file='NSL_KDD/Field Names.csv', with_host=False): selected_parameters = ['duration', 'protocol_type', 'service', 'flag', 'src_bytes', 'dst_bytes', 'land', 'wrong_fragment', 'urgent'] # "Label" - "converter function" dictionary. label_dict = OrderedDict() result = [] with open(labels_file) as lf: labels = csv.reader(lf) for label in labels: if len(label) == 1 or label[1] == 'continuous': label_dict[label[0]] = lambda l: np.float64(l) elif label[1] == 'symbolic': label_dict[label[0]] = lambda l: sh(l) f_list = [i for i in label_dict.values()] n_list = [i for i in label_dict.keys()] data_type = lambda t: t == 'normal' if is_normal else t != 'normal' with open(data_file) as df: # data = csv.DictReader(df, label_dict.keys()) data = csv.reader(df) for d in data: if data_type(d[-2]): # Skip last two fields and add only specified fields. net_params = tuple(f_list[n](i) for n, i in enumerate(d[:-2]) if n_list[n] in selected_parameters) if with_host: host_params = generate_host_activity(is_normal) result.append(net_params + host_params) else: result.append(net_params) hyp.plot(np.array(result), '.', normalize='across', reduce='UMAP', ndims=3, n_clusters=10, animate='spin', palette='viridis', title='Growing Neural Gas on the NSL-KDD [normal={}]'.format(is_normal), # vectorizer='TfidfVectorizer', # precog=False, bullettime=True, chemtrails=True, tail_duration=100, duration=3, rotations=1, legend=False, explore=False, show=True, save_path='./video.mp4') read_ids_data('NSL_KDD/20 Percent Training Set.csv') read_ids_data('NSL_KDD/20 Percent Training Set.csv', is_normal=False)
, " ".
, .
, , :
- .
- , .
, , .
: . , , , .
:
- . , .
- , .
, :
- .
- .
, .
: , .
. , (, , ).
. .
. , IDS.
, ( , 25%).
IDS . NSL-KDD.
:

@startuml start partition { : ; : \n ; } partition { : ; : ; if ( ) then () -[#blue]-> : ; if ( ) then () -[#green]-> : \n .; else () -[#blue]-> endif else () -[#green]-> : \n ; if ( ) then () -[#green]-> : ; else () -[#blue]-> : ; endif endif } stop @enduml
, , .
, .
, , .
, .
, , 20 .
: . , .
— , .
, " " GNG , .
GNG , .
, :
- GNG [18] — , , [17], .
- IGNG [19] — . . . 20 " GNG", , ( [19]).
, , .
: , , .
, GNG ( , ), , .
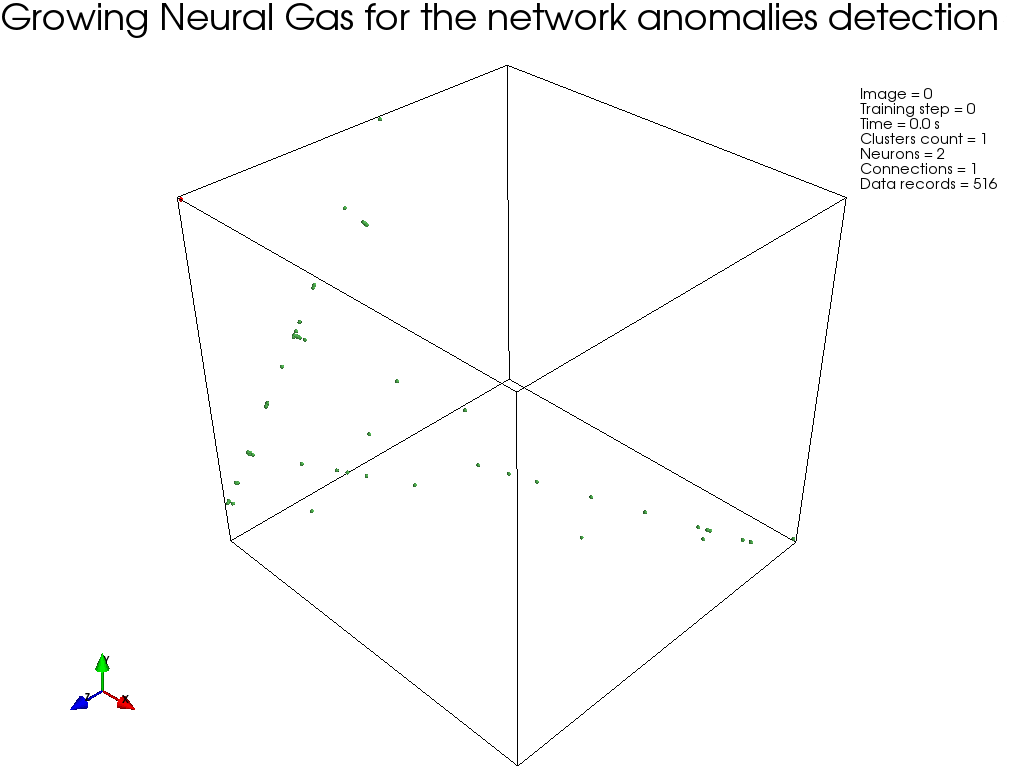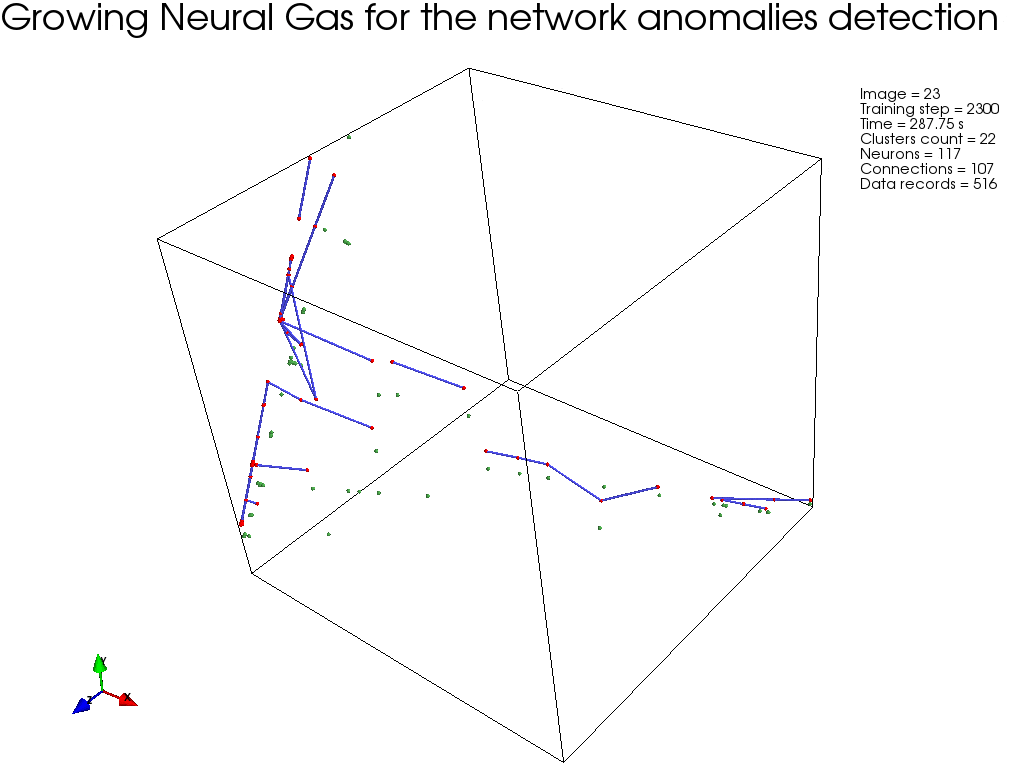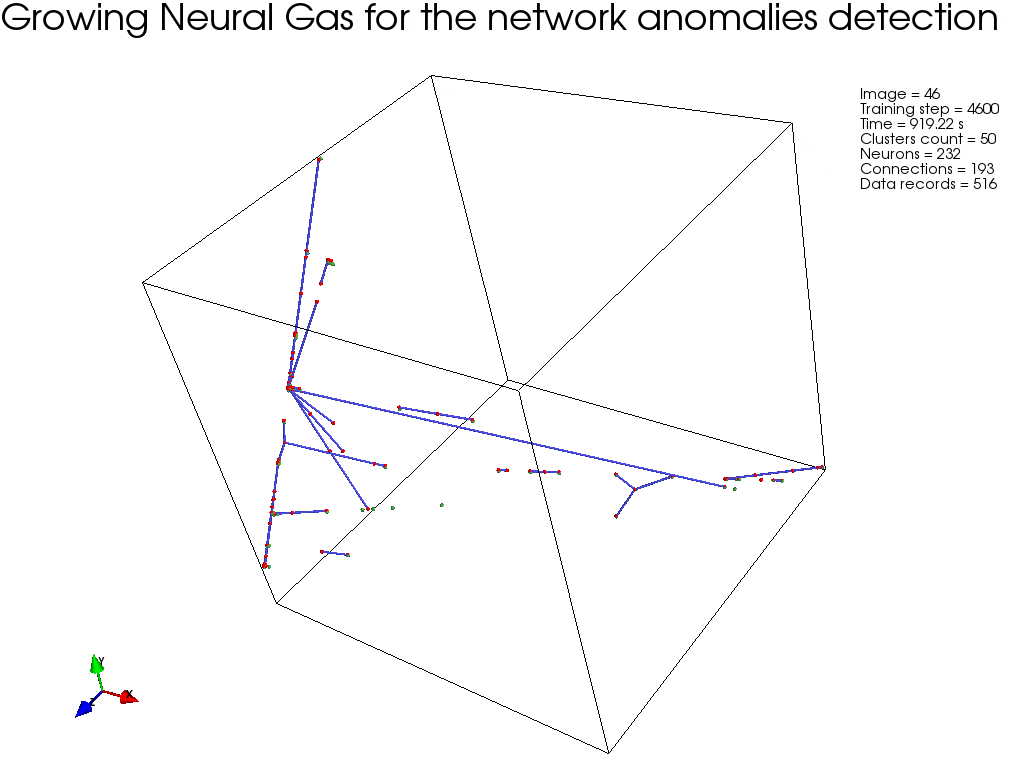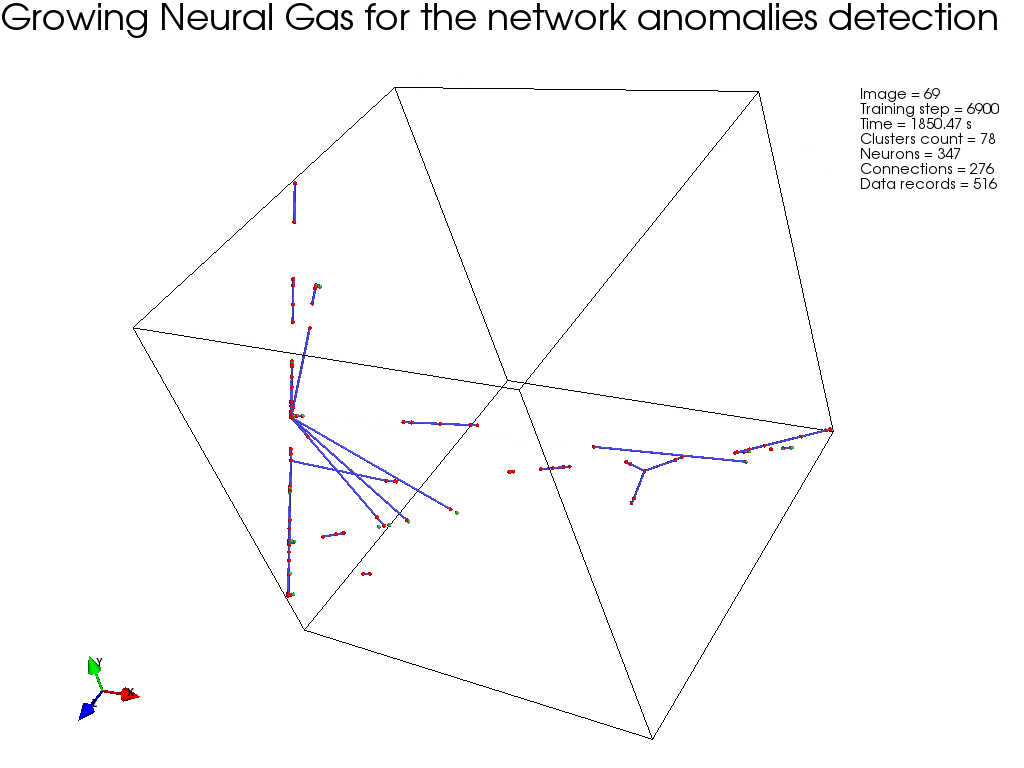
GNG, :
, Python. IGNG , , , .. . , IGNG .
main()
, test_detector()
, , .
test_detector()
.
, , train()
, , detect_anomalies()
.
read_ids_data()
.
NSL-KDD csv
. : , , activity_type
.
, , , generate_host_activity()
, .
Numpy .
, , normalize()
preprocessing
Scikit-Learn . , .
, Graph
NetworkX create_data_graph()
.
.
. , .
GNG
IGNG
, . NeuralGas
, .
train()
__save_img()
.
, .
, (, ) , , .
__save_img()
draw_dots3d()
, draw_graph3d()
, : , .
Mayavi , — mayavi.points3d()
.
, GIF, ImageIO .
:
https://github.com/artiomn/GNG
. , , , (. i
train
: , ). , , IGNG.
( , Github ) .
#!/usr/bin/env python # -*- coding: utf-8 -*- from abc import ABCMeta, abstractmethod from math import sqrt from mayavi import mlab import operator import imageio from collections import OrderedDict from scipy.spatial.distance import euclidean from sklearn import preprocessing import csv import numpy as np import networkx as nx import re import os import shutil import sys import glob from past.builtins import xrange from future.utils import iteritems import time def sh(s): sum = 0 for i, c in enumerate(s): sum += i * ord(c) return sum def create_data_graph(dots): """Create the graph and returns the networkx version of it 'G'.""" count = 0 G = nx.Graph() for i in dots: G.add_node(count, pos=(i)) count += 1 return G def get_ra(ra=0, ra_step=0.3): while True: if ra >= 360: ra = 0 else: ra += ra_step yield ra def shrink_to_3d(data): result = [] for i in data: depth = len(i) if depth <= 3: result.append(i) else: sm = np.sum([(n) * v for n, v in enumerate(i[2:])]) if sm == 0: sm = 1 r = np.array([i[0], i[1], i[2]]) r *= sm r /= np.sum(r) result.append(r) return preprocessing.normalize(result, axis=0, norm='max') def draw_dots3d(dots, edges, fignum, clear=True, title='', size=(1024, 768), graph_colormap='viridis', bgcolor=(1, 1, 1), node_color=(0.3, 0.65, 0.3), node_size=0.01, edge_color=(0.3, 0.3, 0.9), edge_size=0.003, text_size=0.14, text_color=(0, 0, 0), text_coords=[0.84, 0.75], text={}, title_size=0.3, angle=get_ra()): # https://stackoverflow.com/questions/17751552/drawing-multiplex-graphs-with-networkx # numpy array of x, y, z positions in sorted node order xyz = shrink_to_3d(dots) if fignum == 0: mlab.figure(fignum, bgcolor=bgcolor, fgcolor=text_color, size=size) # Mayavi is buggy, and following code causes sockets leak. #if mlab.options.offscreen: # mlab.figure(fignum, bgcolor=bgcolor, fgcolor=text_color, size=size) #elif fignum == 0: # mlab.figure(fignum, bgcolor=bgcolor, fgcolor=text_color, size=size) if clear: mlab.clf() # the x,y, and z co-ordinates are here # manipulate them to obtain the desired projection perspective pts = mlab.points3d(xyz[:, 0], xyz[:, 1], xyz[:, 2], scale_factor=node_size, scale_mode='none', color=node_color, #colormap=graph_colormap, resolution=20, transparent=False) mlab.text(text_coords[0], text_coords[1], '\n'.join(['{} = {}'.format(n, v) for n, v in text.items()]), width=text_size) if clear: mlab.title(title, height=0.95) mlab.roll(next(angle)) mlab.orientation_axes(pts) mlab.outline(pts) """ for i, (x, y, z) in enumerate(xyz): label = mlab.text(x, y, str(i), z=z, width=text_size, name=str(i), color=text_color) label.property.shadow = True """ pts.mlab_source.dataset.lines = edges tube = mlab.pipeline.tube(pts, tube_radius=edge_size) mlab.pipeline.surface(tube, color=edge_color) #mlab.show() # interactive window def draw_graph3d(graph, fignum, *args, **kwargs): graph_pos = nx.get_node_attributes(graph, 'pos') edges = np.array([e for e in graph.edges()]) dots = np.array([graph_pos[v] for v in sorted(graph)], dtype='float64') draw_dots3d(dots, edges, fignum, *args, **kwargs) def generate_host_activity(is_normal): # Host loads is changed only in 25% cases. attack_percent = 25 up_level = (20, 30) # CPU load in percent. cpu_load = (10, 30) # Disk IO per second. iops = (10, 50) # Memory consumption in percent. mem_cons = (30, 60) # Memory consumption in Mb/s. netw_act = (10, 50) cur_up_level = 0 if not is_normal and np.random.randint(0, 100) < attack_percent: cur_up_level = np.random.randint(*up_level) cpu_load = np.random.randint(cur_up_level + cpu_load[0], cur_up_level + cpu_load[1]) iops = np.random.randint(cur_up_level + iops[0], cur_up_level + iops[1]) mem_cons = np.random.randint(cur_up_level + mem_cons[0], cur_up_level + mem_cons[1]) netw_act = np.random.randint(cur_up_level + netw_act[0], cur_up_level + netw_act[1]) return cpu_load, iops, mem_cons, netw_act def read_ids_data(data_file, activity_type='normal', labels_file='NSL_KDD/Field Names.csv', with_host=False): selected_parameters = ['duration', 'protocol_type', 'service', 'flag', 'src_bytes', 'dst_bytes', 'land', 'wrong_fragment', 'urgent', 'serror_rate', 'diff_srv_rate', 'srv_diff_host_rate', 'dst_host_srv_count', 'count'] # "Label" - "converter function" dictionary. label_dict = OrderedDict() result = [] with open(labels_file) as lf: labels = csv.reader(lf) for label in labels: if len(label) == 1 or label[1] == 'continuous': label_dict[label[0]] = lambda l: np.float64(l) elif label[1] == 'symbolic': label_dict[label[0]] = lambda l: sh(l) f_list = [i for i in label_dict.values()] n_list = [i for i in label_dict.keys()] if activity_type == 'normal': data_type = lambda t: t == 'normal' elif activity_type == 'abnormal': data_type = lambda t: t != 'normal' elif activity_type == 'full': data_type = lambda t: True else: raise ValueError('`activity_type` must be "normal", "abnormal" or "full"') print('Reading {} activity from the file "{}" [generated host data {} included]...'. format(activity_type, data_file, 'was' if with_host else 'was not')) with open(data_file) as df: # data = csv.DictReader(df, label_dict.keys()) data = csv.reader(df) for d in data: if data_type(d[-2]): # Skip last two fields and add only specified fields. net_params = tuple(f_list[n](i) for n, i in enumerate(d[:-2]) if n_list[n] in selected_parameters) if with_host: host_params = generate_host_activity(activity_type != 'abnormal') result.append(net_params + host_params) else: result.append(net_params) print('Records count: {}'.format(len(result))) return result class NeuralGas(): __metaclass__ = ABCMeta def __init__(self, data, surface_graph=None, output_images_dir='images'): self._graph = nx.Graph() self._data = data self._surface_graph = surface_graph # Deviation parameters. self._dev_params = None self._output_images_dir = output_images_dir # Nodes count. self._count = 0 if os.path.isdir(output_images_dir): shutil.rmtree('{}'.format(output_images_dir)) print("Ouput images will be saved in: {0}".format(output_images_dir)) os.makedirs(output_images_dir) self._start_time = time.time() @abstractmethod def train(self, max_iterations=100, save_step=0): raise NotImplementedError() def number_of_clusters(self): return nx.number_connected_components(self._graph) def detect_anomalies(self, data, threshold=5, train=False, save_step=100): anomalies_counter, anomaly_records_counter, normal_records_counter = 0, 0, 0 anomaly_level = 0 start_time = self._start_time = time.time() for i, d in enumerate(data): risk_level = self.test_node(d, train) if risk_level != 0: anomaly_records_counter += 1 anomaly_level += risk_level if anomaly_level > threshold: anomalies_counter += 1 #print('Anomaly was detected [count = {}]!'.format(anomalies_counter)) anomaly_level = 0 else: normal_records_counter += 1 if i % save_step == 0: tm = time.time() - start_time print('Abnormal records = {}, Normal records = {}, Detection time = {} s, Time per record = {} s'. format(anomaly_records_counter, normal_records_counter, round(tm, 2), tm / i if i else 0)) tm = time.time() - start_time print('{} [abnormal records = {}, normal records = {}, detection time = {} s, time per record = {} s]'. format('Anomalies were detected (count = {})'.format(anomalies_counter) if anomalies_counter else 'Anomalies weren\'t detected', anomaly_records_counter, normal_records_counter, round(tm, 2), tm / len(data))) return anomalies_counter > 0 def test_node(self, node, train=False): n, dist = self._determine_closest_vertice(node) dev = self._calculate_deviation_params() dev = dev.get(frozenset(nx.node_connected_component(self._graph, n)), dist + 1) dist_sub_dev = dist - dev if dist_sub_dev > 0: return dist_sub_dev if train: self._dev_params = None self._train_on_data_item(node) return 0 @abstractmethod def _train_on_data_item(self, data_item): raise NotImplementedError() @abstractmethod def _save_img(self, fignum, training_step): """.""" raise NotImplementedError() def _calculate_deviation_params(self, distance_function_params={}): if self._dev_params is not None: return self._dev_params clusters = {} dcvd = self._determine_closest_vertice dlen = len(self._data) #dmean = np.mean(self._data, axis=1) #deviation = 0 for node in self._data: n = dcvd(node, **distance_function_params) cluster = clusters.setdefault(frozenset(nx.node_connected_component(self._graph, n[0])), [0, 0]) cluster[0] += n[1] cluster[1] += 1 clusters = {k: sqrt(v[0]/v[1]) for k, v in clusters.items()} self._dev_params = clusters return clusters def _determine_closest_vertice(self, curnode): """.""" pos = nx.get_node_attributes(self._graph, 'pos') kv = zip(*pos.items()) distances = np.linalg.norm(kv[1] - curnode, ord=2, axis=1) i0 = np.argsort(distances)[0] return kv[0][i0], distances[i0] def _determine_2closest_vertices(self, curnode): """Where this curnode is actually the x,y index of the data we want to analyze.""" pos = nx.get_node_attributes(self._graph, 'pos') l_pos = len(pos) if l_pos == 0: return None, None elif l_pos == 1: return pos[0], None kv = zip(*pos.items()) # Calculate Euclidean distance (2-norm of difference vectors) and get first two indexes of the sorted array. # Or a Euclidean-closest nodes index. distances = np.linalg.norm(kv[1] - curnode, ord=2, axis=1) i0, i1 = np.argsort(distances)[0:2] winner1 = tuple((kv[0][i0], distances[i0])) winner2 = tuple((kv[0][i1], distances[i1])) return winner1, winner2 class IGNG(NeuralGas): """Incremental Growing Neural Gas multidimensional implementation""" def __init__(self, data, surface_graph=None, eps_b=0.05, eps_n=0.0005, max_age=10, a_mature=1, output_images_dir='images'): """.""" NeuralGas.__init__(self, data, surface_graph, output_images_dir) self._eps_b = eps_b self._eps_n = eps_n self._max_age = max_age self._a_mature = a_mature self._num_of_input_signals = 0 self._fignum = 0 self._max_train_iters = 0 # Initial value is a standard deviation of the data. self._d = np.std(data) def train(self, max_iterations=100, save_step=0): """IGNG training method""" self._dev_params = None self._max_train_iters = max_iterations fignum = self._fignum self._save_img(fignum, 0) CHS = self.__calinski_harabaz_score igng = self.__igng data = self._data if save_step < 1: save_step = max_iterations old = 0 calin = CHS() i_count = 0 start_time = self._start_time = time.time() while old - calin <= 0: print('Iteration {0:d}...'.format(i_count)) i_count += 1 steps = 1 while steps <= max_iterations: for i, x in enumerate(data): igng(x) if i % save_step == 0: tm = time.time() - start_time print('Training time = {} s, Time per record = {} s, Training step = {}, Clusters count = {}, Neurons = {}, CHI = {}'. format(round(tm, 2), tm / (i if i and i_count == 0 else len(data)), i_count, self.number_of_clusters(), len(self._graph), old - calin) ) self._save_img(fignum, i_count) fignum += 1 steps += 1 self._d -= 0.1 * self._d old = calin calin = CHS() print('Training complete, clusters count = {}, training time = {} s'.format(self.number_of_clusters(), round(time.time() - start_time, 2))) self._fignum = fignum def _train_on_data_item(self, data_item): steps = 0 igng = self.__igng # while steps < self._max_train_iters: while steps < 5: igng(data_item) steps += 1 def __long_train_on_data_item(self, data_item): """.""" np.append(self._data, data_item) self._dev_params = None CHS = self.__calinski_harabaz_score igng = self.__igng data = self._data max_iterations = self._max_train_iters old = 0 calin = CHS() i_count = 0 # Strictly less. while old - calin < 0: print('Training with new normal node, step {0:d}...'.format(i_count)) i_count += 1 steps = 0 if i_count > 100: print('BUG', old, calin) break while steps < max_iterations: igng(data_item) steps += 1 self._d -= 0.1 * self._d old = calin calin = CHS() def _calculate_deviation_params(self, skip_embryo=True): return super(IGNG, self)._calculate_deviation_params(distance_function_params={'skip_embryo': skip_embryo}) def __calinski_harabaz_score(self, skip_embryo=True): graph = self._graph nodes = graph.nodes extra_disp, intra_disp = 0., 0. # CHI = [B / (c - 1)]/[W / (n - c)] # Total numb er of neurons. #ns = nx.get_node_attributes(self._graph, 'n_type') c = len([v for v in nodes.values() if v['n_type'] == 1]) if skip_embryo else len(nodes) # Total number of data. n = len(self._data) # Mean of the all data. mean = np.mean(self._data, axis=1) pos = nx.get_node_attributes(self._graph, 'pos') for node, k in pos.items(): if skip_embryo and nodes[node]['n_type'] == 0: # Skip embryo neurons. continue mean_k = np.mean(k) extra_disp += len(k) * np.sum((mean_k - mean) ** 2) intra_disp += np.sum((k - mean_k) ** 2) return (1. if intra_disp == 0. else extra_disp * (n - c) / (intra_disp * (c - 1.))) def _determine_closest_vertice(self, curnode, skip_embryo=True): """Where this curnode is actually the x,y index of the data we want to analyze.""" pos = nx.get_node_attributes(self._graph, 'pos') nodes = self._graph.nodes distance = sys.maxint for node, position in pos.items(): if skip_embryo and nodes[node]['n_type'] == 0: # Skip embryo neurons. continue dist = euclidean(curnode, position) if dist < distance: distance = dist return node, distance def __get_specific_nodes(self, n_type): return [n for n, p in nx.get_node_attributes(self._graph, 'n_type').items() if p == n_type] def __igng(self, cur_node): """Main IGNG training subroutine""" # find nearest unit and second nearest unit winner1, winner2 = self._determine_2closest_vertices(cur_node) graph = self._graph nodes = graph.nodes d = self._d # Second list element is a distance. if winner1 is None or winner1[1] >= d: # 0 - is an embryo type. graph.add_node(self._count, pos=cur_node, n_type=0, age=0) winner_node1 = self._count self._count += 1 return else: winner_node1 = winner1[0] # Second list element is a distance. if winner2 is None or winner2[1] >= d: # 0 - is an embryo type. graph.add_node(self._count, pos=cur_node, n_type=0, age=0) winner_node2 = self._count self._count += 1 graph.add_edge(winner_node1, winner_node2, age=0) return else: winner_node2 = winner2[0] # Increment the age of all edges, emanating from the winner. for e in graph.edges(winner_node1, data=True): e[2]['age'] += 1 w_node = nodes[winner_node1] # Move the winner node towards current node. w_node['pos'] += self._eps_b * (cur_node - w_node['pos']) neighbors = nx.all_neighbors(graph, winner_node1) a_mature = self._a_mature for n in neighbors: c_node = nodes[n] # Move all direct neighbors of the winner. c_node['pos'] += self._eps_n * (cur_node - c_node['pos']) # Increment the age of all direct neighbors of the winner. c_node['age'] += 1 if c_node['n_type'] == 0 and c_node['age'] >= a_mature: # Now, it's a mature neuron. c_node['n_type'] = 1 # Create connection with age == 0 between two winners. graph.add_edge(winner_node1, winner_node2, age=0) max_age = self._max_age # If there are ages more than maximum allowed age, remove them. age_of_edges = nx.get_edge_attributes(graph, 'age') for edge, age in iteritems(age_of_edges): if age >= max_age: graph.remove_edge(edge[0], edge[1]) # If it causes isolated vertix, remove that vertex as well. #graph.remove_nodes_from(nx.isolates(graph)) for node, v in nodes.items(): if v['n_type'] == 0: # Skip embryo neurons. continue if not graph.neighbors(node): graph.remove_node(node) def _save_img(self, fignum, training_step): """.""" title='Incremental Growing Neural Gas for the network anomalies detection' if self._surface_graph is not None: text = OrderedDict([ ('Image', fignum), ('Training step', training_step), ('Time', '{} s'.format(round(time.time() - self._start_time, 2))), ('Clusters count', self.number_of_clusters()), ('Neurons', len(self._graph)), (' Mature', len(self.__get_specific_nodes(1))), (' Embryo', len(self.__get_specific_nodes(0))), ('Connections', len(self._graph.edges)), ('Data records', len(self._data)) ]) draw_graph3d(self._surface_graph, fignum, title=title) graph = self._graph if len(graph) > 0: #graph_pos = nx.get_node_attributes(graph, 'pos') #nodes = sorted(self.get_specific_nodes(1)) #dots = np.array([graph_pos[v] for v in nodes], dtype='float64') #edges = np.array([e for e in graph.edges(nodes) if e[0] in nodes and e[1] in nodes]) #draw_dots3d(dots, edges, fignum, clear=False, node_color=(1, 0, 0)) draw_graph3d(graph, fignum, clear=False, node_color=(1, 0, 0), title=title, text=text) mlab.savefig("{0}/{1}.png".format(self._output_images_dir, str(fignum))) #mlab.close(fignum) class GNG(NeuralGas): """Growing Neural Gas multidimensional implementation""" def __init__(self, data, surface_graph=None, eps_b=0.05, eps_n=0.0006, max_age=15, lambda_=20, alpha=0.5, d=0.005, max_nodes=1000, output_images_dir='images'): """.""" NeuralGas.__init__(self, data, surface_graph, output_images_dir) self._eps_b = eps_b self._eps_n = eps_n self._max_age = max_age self._lambda = lambda_ self._alpha = alpha self._d = d self._max_nodes = max_nodes self._fignum = 0 self.__add_initial_nodes() def train(self, max_iterations=10000, save_step=50, stop_on_chi=False): """.""" self._dev_params = None self._save_img(self._fignum, 0) graph = self._graph max_nodes = self._max_nodes d = self._d ld = self._lambda alpha = self._alpha update_winner = self.__update_winner data = self._data CHS = self.__calinski_harabaz_score old = 0 calin = CHS() start_time = self._start_time = time.time() train_step = self.__train_step for i in xrange(1, max_iterations): tm = time.time() - start_time print('Training time = {} s, Time per record = {} s, Training step = {}/{}, Clusters count = {}, Neurons = {}'. format(round(tm, 2), tm / len(data), i, max_iterations, self.number_of_clusters(), len(self._graph)) ) for x in data: update_winner(x) train_step(i, alpha, ld, d, max_nodes, True, save_step, graph, update_winner) old = calin calin = CHS() # Stop on the enough clusterization quality. if stop_on_chi and old - calin > 0: break print('Training complete, clusters count = {}, training time = {} s'.format(self.number_of_clusters(), round(time.time() - start_time, 2))) def __train_step(self, i, alpha, ld, d, max_nodes, save_img, save_step, graph, update_winner): g_nodes = graph.nodes # Step 8: if number of input signals generated so far if i % ld == 0 and len(graph) < max_nodes: # Find a node with the largest error. errorvectors = nx.get_node_attributes(graph, 'error') node_largest_error = max(errorvectors.items(), key=operator.itemgetter(1))[0] # Find a node from neighbor of the node just found, with a largest error. neighbors = graph.neighbors(node_largest_error) max_error_neighbor = None max_error = -1 for n in neighbors: ce = g_nodes[n]['error'] if ce > max_error: max_error = ce max_error_neighbor = n # Decrease error variable of other two nodes by multiplying with alpha. new_max_error = alpha * errorvectors[node_largest_error] graph.nodes[node_largest_error]['error'] = new_max_error graph.nodes[max_error_neighbor]['error'] = alpha * max_error # Insert a new unit half way between these two. self._count += 1 new_node = self._count graph.add_node(new_node, pos=self.__get_average_dist(g_nodes[node_largest_error]['pos'], g_nodes[max_error_neighbor]['pos']), error=new_max_error) # Insert edges between new node and other two nodes. graph.add_edge(new_node, max_error_neighbor, age=0) graph.add_edge(new_node, node_largest_error, age=0) # Remove edge between old nodes. graph.remove_edge(max_error_neighbor, node_largest_error) if True and i % save_step == 0: self._fignum += 1 self._save_img(self._fignum, i) # step 9: Decrease all error variables. for n in graph.nodes(): oe = g_nodes[n]['error'] g_nodes[n]['error'] -= d * oe def _train_on_data_item(self, data_item): """IGNG training method""" np.append(self._data, data_item) graph = self._graph max_nodes = self._max_nodes d = self._d ld = self._lambda alpha = self._alpha update_winner = self.__update_winner data = self._data train_step = self.__train_step #for i in xrange(1, 5): update_winner(data_item) train_step(0, alpha, ld, d, max_nodes, False, -1, graph, update_winner) def _calculate_deviation_params(self): return super(GNG, self)._calculate_deviation_params() def __add_initial_nodes(self): """Initialize here""" node1 = self._data[np.random.randint(0, len(self._data))] node2 = self._data[np.random.randint(0, len(self._data))] # make sure you dont select same positions if self.__is_nodes_equal(node1, node2): raise ValueError("Rerun ---------------> similar nodes selected") self._count = 0 self._graph.add_node(self._count, pos=node1, error=0) self._count += 1 self._graph.add_node(self._count, pos=node2, error=0) self._graph.add_edge(self._count - 1, self._count, age=0) def __is_nodes_equal(self, n1, n2): return len(set(n1) & set(n2)) == len(n1) def __update_winner(self, curnode): """.""" # find nearest unit and second nearest unit winner1, winner2 = self._determine_2closest_vertices(curnode) winner_node1 = winner1[0] winner_node2 = winner2[0] win_dist_from_node = winner1[1] graph = self._graph g_nodes = graph.nodes # Update the winner error. g_nodes[winner_node1]['error'] += + win_dist_from_node**2 # Move the winner node towards current node. g_nodes[winner_node1]['pos'] += self._eps_b * (curnode - g_nodes[winner_node1]['pos']) eps_n = self._eps_n # Now update all the neighbors distances. for n in nx.all_neighbors(graph, winner_node1): g_nodes[n]['pos'] += eps_n * (curnode - g_nodes[n]['pos']) # Update age of the edges, emanating from the winner. for e in graph.edges(winner_node1, data=True): e[2]['age'] += 1 # Create or zeroe edge between two winner nodes. graph.add_edge(winner_node1, winner_node2, age=0) # if there are ages more than maximum allowed age, remove them age_of_edges = nx.get_edge_attributes(graph, 'age') max_age = self._max_age for edge, age in age_of_edges.items(): if age >= max_age: graph.remove_edge(edge[0], edge[1]) # If it causes isolated vertix, remove that vertex as well. for node in g_nodes: if not graph.neighbors(node): graph.remove_node(node) def __get_average_dist(self, a, b): """.""" return (a + b) / 2 def __calinski_harabaz_score(self): graph = self._graph nodes = graph.nodes extra_disp, intra_disp = 0., 0. # CHI = [B / (c - 1)]/[W / (n - c)] # Total numb er of neurons. #ns = nx.get_node_attributes(self._graph, 'n_type') c = len(nodes) # Total number of data. n = len(self._data) # Mean of the all data. mean = np.mean(self._data, axis=1) pos = nx.get_node_attributes(self._graph, 'pos') for node, k in pos.items(): mean_k = np.mean(k) extra_disp += len(k) * np.sum((mean_k - mean) ** 2) intra_disp += np.sum((k - mean_k) ** 2) def _save_img(self, fignum, training_step): """.""" title = 'Growing Neural Gas for the network anomalies detection' if self._surface_graph is not None: text = OrderedDict([ ('Image', fignum), ('Training step', training_step), ('Time', '{} s'.format(round(time.time() - self._start_time, 2))), ('Clusters count', self.number_of_clusters()), ('Neurons', len(self._graph)), ('Connections', len(self._graph.edges)), ('Data records', len(self._data)) ]) draw_graph3d(self._surface_graph, fignum, title=title) graph = self._graph if len(graph) > 0: draw_graph3d(graph, fignum, clear=False, node_color=(1, 0, 0), title=title, text=text) mlab.savefig("{0}/{1}.png".format(self._output_images_dir, str(fignum))) def sort_nicely(limages): """Numeric string sort""" def convert(text): return int(text) if text.isdigit() else text def alphanum_key(key): return [convert(c) for c in re.split('([0-9]+)', key)] limages = sorted(limages, key=alphanum_key) return limages def convert_images_to_gif(output_images_dir, output_gif): """Convert a list of images to a gif.""" image_dir = "{0}/*.png".format(output_images_dir) list_images = glob.glob(image_dir) file_names = sort_nicely(list_images) images = [imageio.imread(fn) for fn in file_names] imageio.mimsave(output_gif, images) def test_detector(use_hosts_data, max_iters, alg, output_images_dir='images', output_gif='output.gif'): """Detector quality testing routine""" #data = read_ids_data('NSL_KDD/20 Percent Training Set.csv') frame = '-' * 70 training_set = 'NSL_KDD/Small Training Set.csv' #training_set = 'NSL_KDD/KDDTest-21.txt' testing_set = 'NSL_KDD/KDDTest-21.txt' #testing_set = 'NSL_KDD/KDDTrain+.txt' print('{}\n{}\n{}'.format(frame, '{} detector training...'.format(alg.__name__), frame)) data = read_ids_data(training_set, activity_type='normal', with_host=use_hosts_data) data = preprocessing.normalize(np.array(data, dtype='float64'), axis=1, norm='l1', copy=False) G = create_data_graph(data) gng = alg(data, surface_graph=G, output_images_dir=output_images_dir) gng.train(max_iterations=max_iters, save_step=50) print('Saving GIF file...') convert_images_to_gif(output_images_dir, output_gif) print('{}\n{}\n{}'.format(frame, 'Applying detector to the normal activity using the training set...', frame)) gng.detect_anomalies(data) for a_type in ['abnormal', 'full']: print('{}\n{}\n{}'.format(frame, 'Applying detector to the {} activity using the training set...'.format(a_type), frame)) d_data = read_ids_data(training_set, activity_type=a_type, with_host=use_hosts_data) d_data = preprocessing.normalize(np.array(d_data, dtype='float64'), axis=1, norm='l1', copy=False) gng.detect_anomalies(d_data) dt = OrderedDict([('normal', None), ('abnormal', None), ('full', None)]) for a_type in dt.keys(): print('{}\n{}\n{}'.format(frame, 'Applying detector to the {} activity using the testing set without adaptive learning...'.format(a_type), frame)) d = read_ids_data(testing_set, activity_type=a_type, with_host=use_hosts_data) dt[a_type] = d = preprocessing.normalize(np.array(d, dtype='float64'), axis=1, norm='l1', copy=False) gng.detect_anomalies(d, save_step=1000, train=False) for a_type in ['full']: print('{}\n{}\n{}'.format(frame, 'Applying detector to the {} activity using the testing set with adaptive learning...'.format(a_type), frame)) gng.detect_anomalies(dt[a_type], train=True, save_step=1000) def main(): """Entry point""" start_time = time.time() mlab.options.offscreen = True test_detector(use_hosts_data=False, max_iters=7000, alg=GNG, output_gif='gng_wohosts.gif') print('Working time = {}'.format(round(time.time() - start_time, 2))) test_detector(use_hosts_data=True, max_iters=7000, alg=GNG, output_gif='gng_whosts.gif') print('Working time = {}'.format(round(time.time() - start_time, 2))) test_detector(use_hosts_data=False, max_iters=100, alg=IGNG, output_gif='igng_wohosts.gif') print('Working time = {}'.format(round(time.time() - start_time, 2))) test_detector(use_hosts_data=True, max_iters=100, alg=IGNG, output_gif='igng_whosts.gif') print('Full working time = {}'.format(round(time.time() - start_time, 2))) return 0 if __name__ == "__main__": exit(main())
結果
. , ( ), .
, , .
, .
, , .
, .
:
- GNG: 7000.
- : 516.
- : 495.
- : 2152.
- : 9698.
- : 11850.
---------------------------------------------------------------------- GNG detector training... ---------------------------------------------------------------------- Reading normal activity from the file "NSL_KDD/Small Training Set.csv" [generated host data was not included]... Records count: 516 Ouput images will be saved in: images Training time = 0.0 s, Time per record = 5.54461811864e-09 s, Training step = 1/7000, Clusters count = 1, Neurons = 2 Training time = 0.04 s, Time per record = 7.09027282951e-05 s, Training step = 2/7000, Clusters count = 1, Neurons = 2 Training time = 0.07 s, Time per record = 0.000142523022585 s, Training step = 3/7000, Clusters count = 1, Neurons = 2 Training time = 0.11 s, Time per record = 0.000216007694718 s, Training step = 4/7000, Clusters count = 1, Neurons = 2 Training time = 0.16 s, Time per record = 0.000302943610406 s, Training step = 5/7000, Clusters count = 1, Neurons = 2 Training time = 0.2 s, Time per record = 0.000389085259548 s, Training step = 6/7000, Clusters count = 1, Neurons = 2 Training time = 0.25 s, Time per record = 0.000486224658729 s, Training step = 7/7000, Clusters count = 1, Neurons = 2 Training time = 0.3 s, Time per record = 0.000579897285432 s, Training step = 8/7000, Clusters count = 1, Neurons = 2 Training time = 0.35 s, Time per record = 0.000673654929612 s, Training step = 9/7000, Clusters count = 1, Neurons = 2 ... Training time = 1889.7 s, Time per record = 3.66220002119 s, Training step = 6986/7000, Clusters count = 78, Neurons = 351 Training time = 1890.16 s, Time per record = 3.66309242239 s, Training step = 6987/7000, Clusters count = 78, Neurons = 351 Training time = 1890.61 s, Time per record = 3.6639701858 s, Training step = 6988/7000, Clusters count = 78, Neurons = 351 Training time = 1891.07 s, Time per record = 3.66486349586 s, Training step = 6989/7000, Clusters count = 78, Neurons = 351 Training time = 1891.52 s, Time per record = 3.66574243797 s, Training step = 6990/7000, Clusters count = 78, Neurons = 351 Training time = 1891.98 s, Time per record = 3.66663252291 s, Training step = 6991/7000, Clusters count = 78, Neurons = 351 Training time = 1892.43 s, Time per record = 3.6675082556 s, Training step = 6992/7000, Clusters count = 78, Neurons = 351 Training time = 1892.9 s, Time per record = 3.66840752705 s, Training step = 6993/7000, Clusters count = 78, Neurons = 351 Training time = 1893.35 s, Time per record = 3.66929203087 s, Training step = 6994/7000, Clusters count = 78, Neurons = 351 Training time = 1893.81 s, Time per record = 3.6701756531 s, Training step = 6995/7000, Clusters count = 78, Neurons = 351 Training time = 1894.26 s, Time per record = 3.67105510068 s, Training step = 6996/7000, Clusters count = 78, Neurons = 351 Training time = 1894.71 s, Time per record = 3.67192640508 s, Training step = 6997/7000, Clusters count = 78, Neurons = 351 Training time = 1895.18 s, Time per record = 3.67282555408 s, Training step = 6998/7000, Clusters count = 78, Neurons = 351 Training time = 1895.63 s, Time per record = 3.67370033726 s, Training step = 6999/7000, Clusters count = 78, Neurons = 351 Training complete, clusters count = 78, training time = 1896.09 s Saving GIF file... ---------------------------------------------------------------------- Applying detector to the normal activity using the training set... ---------------------------------------------------------------------- Abnormal records = 0, Normal records = 1, Detection time = 0.2 s, Time per record = 0 s Abnormal records = 0, Normal records = 101, Detection time = 0.24 s, Time per record = 0.00244196891785 s Abnormal records = 0, Normal records = 201, Detection time = 0.28 s, Time per record = 0.00141963005066 s Abnormal records = 0, Normal records = 301, Detection time = 0.32 s, Time per record = 0.00107904990514 s Abnormal records = 0, Normal records = 401, Detection time = 0.36 s, Time per record = 0.000907952189445 s Abnormal records = 0, Normal records = 501, Detection time = 0.4 s, Time per record = 0.000804873943329 s Anomalies weren't detected [abnormal records = 0, normal records = 516, detection time = 0.41 s, time per record = 0.000791241956312 s] ---------------------------------------------------------------------- Applying detector to the abnormal activity using the training set... ---------------------------------------------------------------------- Reading abnormal activity from the file "NSL_KDD/Small Training Set.csv" [generated host data was not included]... Records count: 495 Abnormal records = 0, Normal records = 1, Detection time = 0.0 s, Time per record = 0 s Abnormal records = 62, Normal records = 39, Detection time = 0.04 s, Time per record = 0.000383739471436 s Abnormal records = 133, Normal records = 68, Detection time = 0.08 s, Time per record = 0.000377835035324 s Abnormal records = 198, Normal records = 103, Detection time = 0.12 s, Time per record = 0.000388882954915 s Abnormal records = 269, Normal records = 132, Detection time = 0.15 s, Time per record = 0.000385674834251 s Anomalies were detected (count = 7) [abnormal records = 344, normal records = 151, detection time = 0.19 s, time per record = 0.000390153461032 s] ---------------------------------------------------------------------- Applying detector to the full activity using the training set... ---------------------------------------------------------------------- Reading full activity from the file "NSL_KDD/Small Training Set.csv" [generated host data was not included]... Records count: 1011 Abnormal records = 0, Normal records = 1, Detection time = 0.0 s, Time per record = 0 s Abnormal records = 29, Normal records = 72, Detection time = 0.04 s, Time per record = 0.000397310256958 s Abnormal records = 58, Normal records = 143, Detection time = 0.08 s, Time per record = 0.000394929647446 s Abnormal records = 90, Normal records = 211, Detection time = 0.12 s, Time per record = 0.000392502943675 s Abnormal records = 123, Normal records = 278, Detection time = 0.16 s, Time per record = 0.000393797159195 s Abnormal records = 156, Normal records = 345, Detection time = 0.2 s, Time per record = 0.000392875671387 s Abnormal records = 188, Normal records = 413, Detection time = 0.24 s, Time per record = 0.000391929944356 s Abnormal records = 218, Normal records = 483, Detection time = 0.27 s, Time per record = 0.000391151223864 s Abnormal records = 259, Normal records = 542, Detection time = 0.31 s, Time per record = 0.000390258729458 s Abnormal records = 294, Normal records = 607, Detection time = 0.35 s, Time per record = 0.000389169851939 s Abnormal records = 335, Normal records = 666, Detection time = 0.39 s, Time per record = 0.000388996839523 s Anomalies were detected (count = 7) [abnormal records = 344, normal records = 667, detection time = 0.39 s, time per record = 0.000388572524964 s] ---------------------------------------------------------------------- Applying detector to the normal activity using the testing set without adaptive learning... ---------------------------------------------------------------------- Reading normal activity from the file "NSL_KDD/KDDTest-21.txt" [generated host data was not included]... Records count: 2152 Abnormal records = 1, Normal records = 0, Detection time = 0.0 s, Time per record = 0 s Abnormal records = 368, Normal records = 633, Detection time = 0.4 s, Time per record = 0.000396910905838 s Abnormal records = 737, Normal records = 1264, Detection time = 0.81 s, Time per record = 0.000405857920647 s Anomalies were detected (count = 7) [abnormal records = 794, normal records = 1358, detection time = 0.88 s, time per record = 0.00040672259703 s] ---------------------------------------------------------------------- Applying detector to the abnormal activity using the testing set without adaptive learning... ---------------------------------------------------------------------- Reading abnormal activity from the file "NSL_KDD/KDDTest-21.txt" [generated host data was not included]... Records count: 9698 Abnormal records = 1, Normal records = 0, Detection time = 0.0 s, Time per record = 0 s Abnormal records = 776, Normal records = 225, Detection time = 0.39 s, Time per record = 0.000390892028809 s Abnormal records = 1547, Normal records = 454, Detection time = 0.78 s, Time per record = 0.000388429045677 s Abnormal records = 2332, Normal records = 669, Detection time = 1.16 s, Time per record = 0.000386790037155 s Abnormal records = 3117, Normal records = 884, Detection time = 1.56 s, Time per record = 0.000389873743057 s Abnormal records = 3899, Normal records = 1102, Detection time = 1.95 s, Time per record = 0.000389337825775 s Abnormal records = 4700, Normal records = 1301, Detection time = 2.33 s, Time per record = 0.000388201673826 s Abnormal records = 5502, Normal records = 1499, Detection time = 2.71 s, Time per record = 0.000387295722961 s Abnormal records = 6277, Normal records = 1724, Detection time = 3.1 s, Time per record = 0.000387670874596 s Abnormal records = 7063, Normal records = 1938, Detection time = 3.49 s, Time per record = 0.000387644237942 s Anomalies were detected (count = 154) [abnormal records = 7605, normal records = 2093, detection time = 3.75 s, time per record = 0.000387096114935 s] ---------------------------------------------------------------------- Applying detector to the full activity using the testing set without adaptive learning... ---------------------------------------------------------------------- Reading full activity from the file "NSL_KDD/KDDTest-21.txt" [generated host data was not included]... Records count: 11850 Abnormal records = 1, Normal records = 0, Detection time = 0.0 s, Time per record = 0 s Abnormal records = 698, Normal records = 303, Detection time = 0.39 s, Time per record = 0.000389461040497 s Abnormal records = 1396, Normal records = 605, Detection time = 0.77 s, Time per record = 0.000386721491814 s Abnormal records = 2092, Normal records = 909, Detection time = 1.17 s, Time per record = 0.000389091014862 s Abnormal records = 2808, Normal records = 1193, Detection time = 1.56 s, Time per record = 0.00038931697607 s Abnormal records = 3519, Normal records = 1482, Detection time = 1.95 s, Time per record = 0.000389059782028 s Abnormal records = 4229, Normal records = 1772, Detection time = 2.33 s, Time per record = 0.000388749321302 s Abnormal records = 4957, Normal records = 2044, Detection time = 2.72 s, Time per record = 0.000388323715755 s Abnormal records = 5668, Normal records = 2333, Detection time = 3.11 s, Time per record = 0.00038857537508 s Abnormal records = 6376, Normal records = 2625, Detection time = 3.51 s, Time per record = 0.000389481120639 s Abnormal records = 7079, Normal records = 2922, Detection time = 3.89 s, Time per record = 0.000389060306549 s Abnormal records = 7800, Normal records = 3201, Detection time = 4.27 s, Time per record = 0.000388331630013 s Anomalies were detected (count = 161) [abnormal records = 8399, normal records = 3451, detection time = 4.61 s, time per record = 0.000388800504338 s] Working time = 1914.96 ---------------------------------------------------------------------- GNG detector training... ---------------------------------------------------------------------- Reading normal activity from the file "NSL_KDD/Small Training Set.csv" [generated host data was included]... Records count: 516 Ouput images will be saved in: images Training time = 0.0 s, Time per record = 6.00666962853e-09 s, Training step = 1/7000, Clusters count = 1, Neurons = 2 Training time = 0.04 s, Time per record = 7.04014024069e-05 s, Training step = 2/7000, Clusters count = 1, Neurons = 2 Training time = 0.07 s, Time per record = 0.000143418940463 s, Training step = 3/7000, Clusters count = 1, Neurons = 2 Training time = 0.11 s, Time per record = 0.000213542649912 s, Training step = 4/7000, Clusters count = 1, Neurons = 2 Training time = 0.15 s, Time per record = 0.000284942083581 s, Training step = 5/7000, Clusters count = 1, Neurons = 2 Training time = 0.19 s, Time per record = 0.00037281781204 s, Training step = 6/7000, Clusters count = 1, Neurons = 2 Training time = 0.23 s, Time per record = 0.0004449033922 s, Training step = 7/7000, Clusters count = 1, Neurons = 2 Training time = 0.27 s, Time per record = 0.000518457372059 s, Training step = 8/7000, Clusters count = 1, Neurons = 2 Training time = 0.3 s, Time per record = 0.00058810701666 s, Training step = 9/7000, Clusters count = 1, Neurons = 2 Training time = 0.34 s, Time per record = 0.000659637672957 s, Training step = 10/7000, Clusters count = 1, Neurons = 2 Training time = 0.38 s, Time per record = 0.000728998073312 s, Training step = 11/7000, Clusters count = 1, Neurons = 2 Training time = 0.41 s, Time per record = 0.0007999751919 s, Training step = 12/7000, Clusters count = 1, Neurons = 2 ... Training time = 1832.99 s, Time per record = 3.55230925804 s, Training step = 6987/7000, Clusters count = 82, Neurons = 351 Training time = 1833.46 s, Time per record = 3.55321220973 s, Training step = 6988/7000, Clusters count = 82, Neurons = 351 Training time = 1833.92 s, Time per record = 3.5541091127 s, Training step = 6989/7000, Clusters count = 82, Neurons = 351 Training time = 1834.39 s, Time per record = 3.55502607194 s, Training step = 6990/7000, Clusters count = 82, Neurons = 351 Training time = 1834.85 s, Time per record = 3.55591803882 s, Training step = 6991/7000, Clusters count = 82, Neurons = 351 Training time = 1835.32 s, Time per record = 3.55682536844 s, Training step = 6992/7000, Clusters count = 82, Neurons = 351 Training time = 1835.78 s, Time per record = 3.55772192857 s, Training step = 6993/7000, Clusters count = 82, Neurons = 351 Training time = 1836.26 s, Time per record = 3.55863503107 s, Training step = 6994/7000, Clusters count = 82, Neurons = 351 Training time = 1836.72 s, Time per record = 3.55953075026 s, Training step = 6995/7000, Clusters count = 82, Neurons = 351 Training time = 1837.19 s, Time per record = 3.56043755823 s, Training step = 6996/7000, Clusters count = 82, Neurons = 351 Training time = 1837.65 s, Time per record = 3.56133750012 s, Training step = 6997/7000, Clusters count = 82, Neurons = 351 Training time = 1838.11 s, Time per record = 3.56223588766 s, Training step = 6998/7000, Clusters count = 82, Neurons = 351 Training time = 1838.58 s, Time per record = 3.56314611851 s, Training step = 6999/7000, Clusters count = 82, Neurons = 351 Training complete, clusters count = 82, training time = 1839.05 s Saving GIF file... ---------------------------------------------------------------------- Applying detector to the normal activity using the training set... ---------------------------------------------------------------------- Abnormal records = 0, Normal records = 1, Detection time = 0.22 s, Time per record = 0 s Abnormal records = 0, Normal records = 101, Detection time = 0.26 s, Time per record = 0.00264456033707 s Abnormal records = 0, Normal records = 201, Detection time = 0.31 s, Time per record = 0.00152621984482 s Abnormal records = 0, Normal records = 301, Detection time = 0.35 s, Time per record = 0.00115152041117 s Abnormal records = 0, Normal records = 401, Detection time = 0.39 s, Time per record = 0.000965242385864 s Abnormal records = 0, Normal records = 501, Detection time = 0.43 s, Time per record = 0.000851732254028 s Anomalies weren't detected [abnormal records = 0, normal records = 516, detection time = 0.43 s, time per record = 0.00083744387294 s] ---------------------------------------------------------------------- Applying detector to the abnormal activity using the training set... ---------------------------------------------------------------------- Reading abnormal activity from the file "NSL_KDD/Small Training Set.csv" [generated host data was included]... Records count: 495 Abnormal records = 0, Normal records = 1, Detection time = 0.0 s, Time per record = 0 s Abnormal records = 45, Normal records = 56, Detection time = 0.04 s, Time per record = 0.000407500267029 s Abnormal records = 96, Normal records = 105, Detection time = 0.08 s, Time per record = 0.000401464700699 s Abnormal records = 151, Normal records = 150, Detection time = 0.12 s, Time per record = 0.000398120085398 s Abnormal records = 203, Normal records = 198, Detection time = 0.16 s, Time per record = 0.000406047701836 s Anomalies were detected (count = 5) [abnormal records = 265, normal records = 230, detection time = 0.2 s, time per record = 0.000403349808972 s] ---------------------------------------------------------------------- Applying detector to the full activity using the training set... ---------------------------------------------------------------------- Reading full activity from the file "NSL_KDD/Small Training Set.csv" [generated host data was included]... Records count: 1011 Abnormal records = 0, Normal records = 1, Detection time = 0.0 s, Time per record = 0 s Abnormal records = 20, Normal records = 81, Detection time = 0.04 s, Time per record = 0.00041876077652 s Abnormal records = 50, Normal records = 151, Detection time = 0.08 s, Time per record = 0.000415489673615 s Abnormal records = 76, Normal records = 225, Detection time = 0.13 s, Time per record = 0.000419800281525 s Abnormal records = 101, Normal records = 300, Detection time = 0.17 s, Time per record = 0.000416232347488 s Abnormal records = 125, Normal records = 376, Detection time = 0.21 s, Time per record = 0.000413020133972 s Abnormal records = 155, Normal records = 446, Detection time = 0.25 s, Time per record = 0.000409803390503 s Abnormal records = 190, Normal records = 511, Detection time = 0.29 s, Time per record = 0.000409148420606 s Abnormal records = 220, Normal records = 581, Detection time = 0.33 s, Time per record = 0.000412831306458 s Abnormal records = 253, Normal records = 648, Detection time = 0.37 s, Time per record = 0.000414616531796 s Abnormal records = 289, Normal records = 712, Detection time = 0.41 s, Time per record = 0.000414277076721 s Anomalies were detected (count = 5) [abnormal records = 298, normal records = 713, detection time = 0.42 s, time per record = 0.000413806219129 s] ---------------------------------------------------------------------- Applying detector to the normal activity using the testing set without adaptive learning... ---------------------------------------------------------------------- Reading normal activity from the file "NSL_KDD/KDDTest-21.txt" [generated host data was included]... Records count: 2152 Abnormal records = 1, Normal records = 0, Detection time = 0.0 s, Time per record = 0 s Abnormal records = 324, Normal records = 677, Detection time = 0.4 s, Time per record = 0.000400429010391 s Abnormal records = 646, Normal records = 1355, Detection time = 0.8 s, Time per record = 0.000398404955864 s Anomalies were detected (count = 6) [abnormal records = 695, normal records = 1457, detection time = 0.86 s, time per record = 0.000401378675021 s] ---------------------------------------------------------------------- Applying detector to the abnormal activity using the testing set without adaptive learning... ---------------------------------------------------------------------- Reading abnormal activity from the file "NSL_KDD/KDDTest-21.txt" [generated host data was included]... Records count: 9698 Abnormal records = 1, Normal records = 0, Detection time = 0.0 s, Time per record = 0 s Abnormal records = 716, Normal records = 285, Detection time = 0.39 s, Time per record = 0.000391721010208 s Abnormal records = 1424, Normal records = 577, Detection time = 0.79 s, Time per record = 0.000395292520523 s Abnormal records = 2144, Normal records = 857, Detection time = 1.18 s, Time per record = 0.000393549998601 s Abnormal records = 2877, Normal records = 1124, Detection time = 1.57 s, Time per record = 0.000393224000931 s Abnormal records = 3591, Normal records = 1410, Detection time = 1.97 s, Time per record = 0.000394224214554 s Abnormal records = 4337, Normal records = 1664, Detection time = 2.36 s, Time per record = 0.000393829345703 s Abnormal records = 5089, Normal records = 1912, Detection time = 2.77 s, Time per record = 0.000395649433136 s Abnormal records = 5798, Normal records = 2203, Detection time = 3.16 s, Time per record = 0.000395138502121 s Abnormal records = 6532, Normal records = 2469, Detection time = 3.55 s, Time per record = 0.000394880347782 s Anomalies were detected (count = 131) [abnormal records = 7027, normal records = 2671, detection time = 3.84 s, time per record = 0.000396096625262 s] ---------------------------------------------------------------------- Applying detector to the full activity using the testing set without adaptive learning... ---------------------------------------------------------------------- Reading full activity from the file "NSL_KDD/KDDTest-21.txt" [generated host data was included]... Records count: 11850 Abnormal records = 1, Normal records = 0, Detection time = 0.0 s, Time per record = 0 s Abnormal records = 622, Normal records = 379, Detection time = 0.4 s, Time per record = 0.000395487070084 s Abnormal records = 1251, Normal records = 750, Detection time = 0.79 s, Time per record = 0.000395890474319 s Abnormal records = 1876, Normal records = 1125, Detection time = 1.19 s, Time per record = 0.000395860671997 s Abnormal records = 2528, Normal records = 1473, Detection time = 1.6 s, Time per record = 0.000399308979511 s Abnormal records = 3184, Normal records = 1817, Detection time = 1.99 s, Time per record = 0.000397848176956 s Abnormal records = 3824, Normal records = 2177, Detection time = 2.39 s, Time per record = 0.000397575179736 s Abnormal records = 4498, Normal records = 2503, Detection time = 2.79 s, Time per record = 0.000398371560233 s Abnormal records = 5145, Normal records = 2856, Detection time = 3.18 s, Time per record = 0.00039776262641 s Abnormal records = 5792, Normal records = 3209, Detection time = 3.59 s, Time per record = 0.000398459778892 s Abnormal records = 6424, Normal records = 3577, Detection time = 3.98 s, Time per record = 0.000397949695587 s Abnormal records = 7080, Normal records = 3921, Detection time = 4.38 s, Time per record = 0.000397751092911 s Anomalies were detected (count = 138) [abnormal records = 7619, normal records = 4231, detection time = 4.72 s, time per record = 0.000398021931387 s] Working time = 3772.93 ---------------------------------------------------------------------- IGNG detector training... ---------------------------------------------------------------------- Reading normal activity from the file "NSL_KDD/Small Training Set.csv" [generated host data was not included]... Records count: 516 Ouput images will be saved in: images Iteration 0... Training time = 0.0 s, Time per record = 4.85154085381e-08 s, Training step = 1, Clusters count = 1, Neurons = 1, CHI = -1.0 Training time = 0.48 s, Time per record = 0.000924686128779 s, Training step = 1, Clusters count = 11, Neurons = 22, CHI = -1.0 Training time = 0.96 s, Time per record = 0.00185829262401 s, Training step = 1, Clusters count = 10, Neurons = 28, CHI = -1.0 Training time = 1.45 s, Time per record = 0.00280647129976 s, Training step = 1, Clusters count = 11, Neurons = 31, CHI = -1.0 Training time = 1.93 s, Time per record = 0.0037418566933 s, Training step = 1, Clusters count = 11, Neurons = 33, CHI = -1.0 Training time = 2.42 s, Time per record = 0.00468139177145 s, Training step = 1, Clusters count = 11, Neurons = 36, CHI = -1.0 Training time = 2.9 s, Time per record = 0.00562962635543 s, Training step = 1, Clusters count = 14, Neurons = 40, CHI = -1.0 ... Training time = 542.57 s, Time per record = 1.05149506783 s, Training step = 1, Clusters count = 20, Neurons = 65, CHI = -1.0 Training time = 543.07 s, Time per record = 1.05246509692 s, Training step = 1, Clusters count = 21, Neurons = 65, CHI = -1.0 Training time = 543.57 s, Time per record = 1.05343477717 s, Training step = 1, Clusters count = 21, Neurons = 65, CHI = -1.0 Training time = 544.07 s, Time per record = 1.05440813395 s, Training step = 1, Clusters count = 21, Neurons = 65, CHI = -1.0 Training time = 544.59 s, Time per record = 1.05540252148 s, Training step = 1, Clusters count = 19, Neurons = 65, CHI = -1.0 Training time = 545.08 s, Time per record = 1.05635387398 s, Training step = 1, Clusters count = 20, Neurons = 65, CHI = -1.0 Training time = 545.58 s, Time per record = 1.05732676179 s, Training step = 1, Clusters count = 21, Neurons = 65, CHI = -1.0 Training time = 546.22 s, Time per record = 1.05855851395 s, Training step = 1, Clusters count = 21, Neurons = 65, CHI = -1.0 Training time = 546.72 s, Time per record = 1.0595340641 s, Training step = 1, Clusters count = 21, Neurons = 65, CHI = -1.0 Training time = 547.22 s, Time per record = 1.06049638571 s, Training step = 1, Clusters count = 20, Neurons = 65, CHI = -1.0 Training time = 547.71 s, Time per record = 1.06145610689 s, Training step = 1, Clusters count = 20, Neurons = 65, CHI = -1.0 Training time = 548.21 s, Time per record = 1.06243183955 s, Training step = 1, Clusters count = 20, Neurons = 65, CHI = -1.0 Training time = 548.7 s, Time per record = 1.06338078292 s, Training step = 1, Clusters count = 21, Neurons = 65, CHI = -1.0 Training time = 549.2 s, Time per record = 1.0643380252 s, Training step = 1, Clusters count = 21, Neurons = 65, CHI = -1.0 Training time = 549.7 s, Time per record = 1.06530855654 s, Training step = 1, Clusters count = 21, Neurons = 65, CHI = -1.0 Training time = 550.19 s, Time per record = 1.0662655932 s, Training step = 1, Clusters count = 19, Neurons = 65, CHI = -1.0 Training time = 550.69 s, Time per record = 1.06722681606 s, Training step = 1, Clusters count = 20, Neurons = 65, CHI = -1.0 Training time = 551.19 s, Time per record = 1.06818968427 s, Training step = 1, Clusters count = 21, Neurons = 65, CHI = -1.0 Training complete, clusters count = 21, training time = 551.68 s Saving GIF file... ---------------------------------------------------------------------- Applying detector to the normal activity using the training set... ---------------------------------------------------------------------- Abnormal records = 0, Normal records = 1, Detection time = 0.53 s, Time per record = 0 s Abnormal records = 0, Normal records = 101, Detection time = 0.63 s, Time per record = 0.00633862018585 s Abnormal records = 0, Normal records = 201, Detection time = 0.73 s, Time per record = 0.00366881489754 s Abnormal records = 0, Normal records = 301, Detection time = 0.83 s, Time per record = 0.00277556975683 s Abnormal records = 0, Normal records = 401, Detection time = 0.93 s, Time per record = 0.00232824504375 s Abnormal records = 0, Normal records = 501, Detection time = 1.03 s, Time per record = 0.00206651210785 s Anomalies weren't detected [abnormal records = 0, normal records = 516, detection time = 1.05 s, time per record = 0.00203575598177 s] ---------------------------------------------------------------------- Applying detector to the abnormal activity using the training set... ---------------------------------------------------------------------- Reading abnormal activity from the file "NSL_KDD/Small Training Set.csv" [generated host data was not included]... Records count: 495 Abnormal records = 0, Normal records = 1, Detection time = 0.0 s, Time per record = 0 s Abnormal records = 19, Normal records = 82, Detection time = 0.11 s, Time per record = 0.00108736038208 s Abnormal records = 41, Normal records = 160, Detection time = 0.21 s, Time per record = 0.00104064464569 s Abnormal records = 59, Normal records = 242, Detection time = 0.31 s, Time per record = 0.00102363348007 s Abnormal records = 78, Normal records = 323, Detection time = 0.41 s, Time per record = 0.00101397752762 s Anomalies were detected (count = 1) [abnormal records = 110, normal records = 385, detection time = 0.5 s, time per record = 0.00100804242221 s] ---------------------------------------------------------------------- Applying detector to the full activity using the training set... ---------------------------------------------------------------------- Reading full activity from the file "NSL_KDD/Small Training Set.csv" [generated host data was not included]... Records count: 1011 Abnormal records = 0, Normal records = 1, Detection time = 0.0 s, Time per record = 0 s Abnormal records = 6, Normal records = 95, Detection time = 0.1 s, Time per record = 0.00101613998413 s Abnormal records = 18, Normal records = 183, Detection time = 0.2 s, Time per record = 0.00100832462311 s Abnormal records = 30, Normal records = 271, Detection time = 0.3 s, Time per record = 0.00100745360057 s Abnormal records = 38, Normal records = 363, Detection time = 0.41 s, Time per record = 0.00101710498333 s Abnormal records = 48, Normal records = 453, Detection time = 0.51 s, Time per record = 0.00101808595657 s Abnormal records = 58, Normal records = 543, Detection time = 0.61 s, Time per record = 0.00101492325465 s Abnormal records = 68, Normal records = 633, Detection time = 0.71 s, Time per record = 0.00101190873555 s Abnormal records = 76, Normal records = 725, Detection time = 0.81 s, Time per record = 0.00101043373346 s Abnormal records = 88, Normal records = 813, Detection time = 0.91 s, Time per record = 0.00100836992264 s Abnormal records = 105, Normal records = 896, Detection time = 1.01 s, Time per record = 0.00100904512405 s Anomalies were detected (count = 1) [abnormal records = 110, normal records = 901, detection time = 1.02 s, time per record = 0.00100898412521 s] ---------------------------------------------------------------------- Applying detector to the normal activity using the testing set without adaptive learning... ---------------------------------------------------------------------- Reading normal activity from the file "NSL_KDD/KDDTest-21.txt" [generated host data was not included]... Records count: 2152 Abnormal records = 1, Normal records = 0, Detection time = 0.0 s, Time per record = 0 s Abnormal records = 165, Normal records = 836, Detection time = 1.01 s, Time per record = 0.00101356911659 s Abnormal records = 344, Normal records = 1657, Detection time = 2.04 s, Time per record = 0.00101889550686 s Anomalies weren't detected [abnormal records = 372, normal records = 1780, detection time = 2.19 s, time per record = 0.00101830303447 s] ---------------------------------------------------------------------- Applying detector to the abnormal activity using the testing set without adaptive learning... ---------------------------------------------------------------------- Reading abnormal activity from the file "NSL_KDD/KDDTest-21.txt" [generated host data was not included]... Records count: 9698 Abnormal records = 0, Normal records = 1, Detection time = 0.0 s, Time per record = 0 s Abnormal records = 289, Normal records = 712, Detection time = 1.01 s, Time per record = 0.0010106010437 s Abnormal records = 531, Normal records = 1470, Detection time = 2.03 s, Time per record = 0.00101473748684 s Abnormal records = 806, Normal records = 2195, Detection time = 3.05 s, Time per record = 0.00101690498988 s Abnormal records = 1085, Normal records = 2916, Detection time = 4.07 s, Time per record = 0.00101814824343 s Abnormal records = 1340, Normal records = 3661, Detection time = 5.1 s, Time per record = 0.00102022538185 s Abnormal records = 1615, Normal records = 4386, Detection time = 6.11 s, Time per record = 0.00101814981302 s Abnormal records = 1910, Normal records = 5091, Detection time = 7.12 s, Time per record = 0.00101696658134 s Abnormal records = 2177, Normal records = 5824, Detection time = 8.13 s, Time per record = 0.00101655423641 s Abnormal records = 2437, Normal records = 6564, Detection time = 9.14 s, Time per record = 0.00101562854979 s Anomalies were detected (count = 27) [abnormal records = 2602, normal records = 7096, detection time = 9.84 s, time per record = 0.00101498390724 s] ---------------------------------------------------------------------- Applying detector to the full activity using the testing set without adaptive learning... ---------------------------------------------------------------------- Reading full activity from the file "NSL_KDD/KDDTest-21.txt" [generated host data was not included]... Records count: 11850 Abnormal records = 0, Normal records = 1, Detection time = 0.0 s, Time per record = 0 s Abnormal records = 264, Normal records = 737, Detection time = 1.01 s, Time per record = 0.00101314496994 s Abnormal records = 496, Normal records = 1505, Detection time = 2.04 s, Time per record = 0.00101831400394 s Abnormal records = 748, Normal records = 2253, Detection time = 3.07 s, Time per record = 0.0010246480306 s Abnormal records = 1000, Normal records = 3001, Detection time = 4.08 s, Time per record = 0.00101987397671 s Abnormal records = 1264, Normal records = 3737, Detection time = 5.09 s, Time per record = 0.00101791639328 s Abnormal records = 1488, Normal records = 4513, Detection time = 6.1 s, Time per record = 0.00101627349854 s Abnormal records = 1761, Normal records = 5240, Detection time = 7.11 s, Time per record = 0.00101523229054 s Abnormal records = 2019, Normal records = 5982, Detection time = 8.11 s, Time per record = 0.00101378926635 s Abnormal records = 2282, Normal records = 6719, Detection time = 9.11 s, Time per record = 0.00101193467776 s Abnormal records = 2530, Normal records = 7471, Detection time = 10.11 s, Time per record = 0.00101094179153 s Abnormal records = 2781, Normal records = 8220, Detection time = 11.12 s, Time per record = 0.00101091491092 s Anomalies were detected (count = 28) [abnormal records = 2974, normal records = 8876, detection time = 11.97 s, time per record = 0.00101001697251 s] Working time = 5537.67 ---------------------------------------------------------------------- IGNG detector training... ---------------------------------------------------------------------- Reading normal activity from the file "NSL_KDD/Small Training Set.csv" [generated host data was included]... Records count: 516 Ouput images will be saved in: images Iteration 0... Training time = 0.0 s, Time per record = 5.03636145777e-08 s, Training step = 1, Clusters count = 1, Neurons = 1, CHI = -1.0 Training time = 0.5 s, Time per record = 0.000959633856781 s, Training step = 1, Clusters count = 11, Neurons = 24, CHI = -1.0 Training time = 1.0 s, Time per record = 0.00193044566369 s, Training step = 1, Clusters count = 11, Neurons = 32, CHI = -1.0 Training time = 1.5 s, Time per record = 0.00290305069251 s, Training step = 1, Clusters count = 13, Neurons = 38, CHI = -1.0 Training time = 2.0 s, Time per record = 0.0038706590963 s, Training step = 1, Clusters count = 16, Neurons = 40, CHI = -1.0 Training time = 2.5 s, Time per record = 0.00484266669251 s, Training step = 1, Clusters count = 14, Neurons = 42, CHI = -1.0 Training time = 3.0 s, Time per record = 0.00581014156342 s, Training step = 1, Clusters count = 17, Neurons = 46, CHI = -1.0 ... Training time = 555.39 s, Time per record = 1.07633200958 s, Training step = 1, Clusters count = 22, Neurons = 70, CHI = -1.0 Training time = 555.9 s, Time per record = 1.07732224233 s, Training step = 1, Clusters count = 22, Neurons = 70, CHI = -1.0 Training time = 556.41 s, Time per record = 1.07831773093 s, Training step = 1, Clusters count = 22, Neurons = 70, CHI = -1.0 Training time = 556.93 s, Time per record = 1.07931286651 s, Training step = 1, Clusters count = 22, Neurons = 70, CHI = -1.0 Training time = 557.44 s, Time per record = 1.08031143421 s, Training step = 1, Clusters count = 23, Neurons = 70, CHI = -1.0 Training time = 557.95 s, Time per record = 1.08129599806 s, Training step = 1, Clusters count = 23, Neurons = 70, CHI = -1.0 Training time = 558.47 s, Time per record = 1.08229697305 s, Training step = 1, Clusters count = 23, Neurons = 70, CHI = -1.0 Training time = 558.98 s, Time per record = 1.08329910687 s, Training step = 1, Clusters count = 23, Neurons = 70, CHI = -1.0 Training time = 559.51 s, Time per record = 1.08431409496 s, Training step = 1, Clusters count = 22, Neurons = 70, CHI = -1.0 Training time = 560.02 s, Time per record = 1.08530911826 s, Training step = 1, Clusters count = 22, Neurons = 70, CHI = -1.0 Training time = 560.53 s, Time per record = 1.0863001707 s, Training step = 1, Clusters count = 22, Neurons = 70, CHI = -1.0 Training time = 561.05 s, Time per record = 1.08729686062 s, Training step = 1, Clusters count = 22, Neurons = 70, CHI = -1.0 Training time = 561.57 s, Time per record = 1.08831085854 s, Training step = 1, Clusters count = 22, Neurons = 70, CHI = -1.0 Training time = 562.07 s, Time per record = 1.08928472682 s, Training step = 1, Clusters count = 22, Neurons = 70, CHI = -1.0 Training time = 562.58 s, Time per record = 1.09027189525 s, Training step = 1, Clusters count = 22, Neurons = 70, CHI = -1.0 Training time = 563.09 s, Time per record = 1.09126116693 s, Training step = 1, Clusters count = 23, Neurons = 70, CHI = -1.0 Training time = 563.61 s, Time per record = 1.09226539523 s, Training step = 1, Clusters count = 23, Neurons = 70, CHI = -1.0 Training time = 564.13 s, Time per record = 1.09326583933 s, Training step = 1, Clusters count = 23, Neurons = 70, CHI = -1.0 Training time = 564.64 s, Time per record = 1.09426376385 s, Training step = 1, Clusters count = 23, Neurons = 70, CHI = -1.0 Training time = 565.15 s, Time per record = 1.09525539755 s, Training step = 1, Clusters count = 22, Neurons = 70, CHI = -1.0 Training time = 565.67 s, Time per record = 1.09626085259 s, Training step = 1, Clusters count = 22, Neurons = 70, CHI = -1.0 Training time = 566.2 s, Time per record = 1.09728375193 s, Training step = 1, Clusters count = 22, Neurons = 70, CHI = -1.0 Training time = 566.72 s, Time per record = 1.09829355996 s, Training step = 1, Clusters count = 22, Neurons = 70, CHI = -1.0 Training time = 567.24 s, Time per record = 1.09930341697 s, Training step = 1, Clusters count = 22, Neurons = 70, CHI = -1.0 Training complete, clusters count = 22, training time = 567.74 s Saving GIF file... ---------------------------------------------------------------------- Applying detector to the normal activity using the training set... ---------------------------------------------------------------------- Abnormal records = 0, Normal records = 1, Detection time = 0.58 s, Time per record = 0 s Abnormal records = 0, Normal records = 101, Detection time = 0.69 s, Time per record = 0.00693108081818 s Abnormal records = 0, Normal records = 201, Detection time = 0.81 s, Time per record = 0.00403473496437 s Abnormal records = 0, Normal records = 301, Detection time = 0.92 s, Time per record = 0.00306363026301 s Abnormal records = 0, Normal records = 401, Detection time = 1.03 s, Time per record = 0.00257846474648 s Abnormal records = 0, Normal records = 501, Detection time = 1.15 s, Time per record = 0.00229543209076 s Anomalies weren't detected [abnormal records = 0, normal records = 516, detection time = 1.16 s, time per record = 0.00225632181463 s] ---------------------------------------------------------------------- Applying detector to the abnormal activity using the training set... ---------------------------------------------------------------------- Reading abnormal activity from the file "NSL_KDD/Small Training Set.csv" [generated host data was included]... Records count: 495 Abnormal records = 0, Normal records = 1, Detection time = 0.0 s, Time per record = 0 s Abnormal records = 19, Normal records = 82, Detection time = 0.11 s, Time per record = 0.00114132165909 s Abnormal records = 42, Normal records = 159, Detection time = 0.23 s, Time per record = 0.00113487005234 s Abnormal records = 59, Normal records = 242, Detection time = 0.34 s, Time per record = 0.00112538019816 s Abnormal records = 79, Normal records = 322, Detection time = 0.45 s, Time per record = 0.00113292753696 s Anomalies were detected (count = 1) [abnormal records = 111, normal records = 384, detection time = 0.56 s, time per record = 0.0011294605756 s] ---------------------------------------------------------------------- Applying detector to the full activity using the training set... ---------------------------------------------------------------------- Reading full activity from the file "NSL_KDD/Small Training Set.csv" [generated host data was included]... Records count: 1011 Abnormal records = 0, Normal records = 1, Detection time = 0.0 s, Time per record = 0 s Abnormal records = 6, Normal records = 95, Detection time = 0.11 s, Time per record = 0.00112770080566 s Abnormal records = 18, Normal records = 183, Detection time = 0.23 s, Time per record = 0.00115751504898 s Abnormal records = 30, Normal records = 271, Detection time = 0.34 s, Time per record = 0.00114720344543 s Abnormal records = 38, Normal records = 363, Detection time = 0.45 s, Time per record = 0.00113685250282 s Abnormal records = 47, Normal records = 454, Detection time = 0.57 s, Time per record = 0.00113049221039 s Abnormal records = 57, Normal records = 544, Detection time = 0.68 s, Time per record = 0.00112626830737 s Abnormal records = 67, Normal records = 634, Detection time = 0.79 s, Time per record = 0.00113069295883 s Abnormal records = 75, Normal records = 726, Detection time = 0.9 s, Time per record = 0.00112736135721 s Abnormal records = 87, Normal records = 814, Detection time = 1.01 s, Time per record = 0.00112713442908 s Abnormal records = 104, Normal records = 897, Detection time = 1.13 s, Time per record = 0.00112596797943 s Anomalies were detected (count = 1) [abnormal records = 109, normal records = 902, detection time = 1.14 s, time per record = 0.0011247270303 s] ---------------------------------------------------------------------- Applying detector to the normal activity using the testing set without adaptive learning... ---------------------------------------------------------------------- Reading normal activity from the file "NSL_KDD/KDDTest-21.txt" [generated host data was included]... Records count: 2152 Abnormal records = 1, Normal records = 0, Detection time = 0.0 s, Time per record = 0 s Abnormal records = 165, Normal records = 836, Detection time = 1.13 s, Time per record = 0.00113072776794 s Abnormal records = 343, Normal records = 1658, Detection time = 2.25 s, Time per record = 0.00112573599815 s Anomalies weren't detected [abnormal records = 371, normal records = 1781, detection time = 2.42 s, time per record = 0.00112456171929 s] ---------------------------------------------------------------------- Applying detector to the abnormal activity using the testing set without adaptive learning... ---------------------------------------------------------------------- Reading abnormal activity from the file "NSL_KDD/KDDTest-21.txt" [generated host data was included]... Records count: 9698 Abnormal records = 0, Normal records = 1, Detection time = 0.0 s, Time per record = 0 s Abnormal records = 281, Normal records = 720, Detection time = 1.14 s, Time per record = 0.0011370780468 s Abnormal records = 514, Normal records = 1487, Detection time = 2.27 s, Time per record = 0.00113358747959 s Abnormal records = 785, Normal records = 2216, Detection time = 3.39 s, Time per record = 0.00112900535266 s Abnormal records = 1057, Normal records = 2944, Detection time = 4.51 s, Time per record = 0.00112772023678 s Abnormal records = 1297, Normal records = 3704, Detection time = 5.63 s, Time per record = 0.00112658400536 s Abnormal records = 1568, Normal records = 4433, Detection time = 6.76 s, Time per record = 0.00112644219398 s Abnormal records = 1850, Normal records = 5151, Detection time = 7.89 s, Time per record = 0.00112774443626 s Abnormal records = 2105, Normal records = 5896, Detection time = 9.02 s, Time per record = 0.00112706288695 s Abnormal records = 2357, Normal records = 6644, Detection time = 10.14 s, Time per record = 0.00112684233983 s Anomalies were detected (count = 22) [abnormal records = 2521, normal records = 7177, detection time = 10.92 s, time per record = 0.00112578553695 s] ---------------------------------------------------------------------- Applying detector to the full activity using the testing set without adaptive learning... ---------------------------------------------------------------------- Reading full activity from the file "NSL_KDD/KDDTest-21.txt" [generated host data was included]... Records count: 11850 Abnormal records = 0, Normal records = 1, Detection time = 0.0 s, Time per record = 0 s Abnormal records = 253, Normal records = 748, Detection time = 1.13 s, Time per record = 0.00112664413452 s Abnormal records = 479, Normal records = 1522, Detection time = 2.26 s, Time per record = 0.00113046598434 s Abnormal records = 722, Normal records = 2279, Detection time = 3.38 s, Time per record = 0.00112577366829 s Abnormal records = 971, Normal records = 3030, Detection time = 4.51 s, Time per record = 0.00112768054008 s Abnormal records = 1225, Normal records = 3776, Detection time = 5.64 s, Time per record = 0.00112878522873 s Abnormal records = 1439, Normal records = 4562, Detection time = 6.76 s, Time per record = 0.00112742733955 s Abnormal records = 1707, Normal records = 5294, Detection time = 7.89 s, Time per record = 0.00112738772801 s Abnormal records = 1956, Normal records = 6045, Detection time = 9.01 s, Time per record = 0.00112660801411 s Abnormal records = 2205, Normal records = 6796, Detection time = 10.14 s, Time per record = 0.00112647589048 s Abnormal records = 2441, Normal records = 7560, Detection time = 11.26 s, Time per record = 0.0011262802124 s Abnormal records = 2685, Normal records = 8316, Detection time = 12.39 s, Time per record = 0.00112592681971 s Anomalies were detected (count = 23) [abnormal records = 2878, normal records = 8972, detection time = 13.34 s, time per record = 0.00112551166035 s] Full working time = 6245.14
:
-
l_time
— . -
te_l_time
— , . -
te_t_time
— . -
g_l_perc
— , . -
g_t_perc
— . -
f_l_perc
— , . -
f_t_perc
— .
| 種類 | l_time | te_l_time | te_t_time | g_l_perc | g_t_perc | f_l_perc | f_t_perc |
|---|---|---|---|---|---|---|---|
| GNG host | 1869 | 0.39 | 4.61 | 69.5 | 78.4 | 0 | 36.9 |
| GNG host | 1839 | 0.42 | 4.72 | 60.2 | 72.5 | 0 | 47.7 |
| IGNG host | 551 | 1.02 | 11.97 | 22.2 | 26.8 | 0 | 17.3 |
| IGNG host | 567 | 1.14 | 13.34 | 28.9 | 27.0 | 0 | 20.8 |
結論:
- .
- .
- IGNG . , , , (, ).
- , GNG , .. 1.5-2 .
おわりに
, . , . .
- Fast GNG .
- " " [21] [18].
- SOINN , GNG, .
- , .
- .
,
- .
- ハニーポットでのトレーニング。ハニーポットとの相互作用は攻撃と見なすことができ、検出器のトレーニングに使用できます。
- トレーニングパラメータの選択の自動化。このトピックは別の記事を対象としていますが、この例では、適用してみると面白いでしょう。たとえば、TPOTを使用します。遺伝的アルゴリズムをパラメーターに適用すると、それらを手動で選択するよりも迅速かつ正確に選択できるようになります。
コメント、質問、説明を楽しみにしています。
[1] http://www.irjcs.com/volumes/vol4/iss09/08.SISPCS10095.pdf "J.Rubina Parveen — Neural networks in cyber security — 2017"
[2] http://e-notabene.ru/nb/article_18834.html " .. — — 2016"
[3] https://www.sciencedirect.com/science/article/pii/S2352864816300281 "Haibo Zhang, Qing Huang, Fangwei Li, Jiang Zhu — A network security situation prediction model based on wavelet neural network with optimized parameters — 2016"
[4] https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4896428/pdf/pone.0155781.pdf "Min-Joo Kang, Je-Won Kang — Intrusion Detection System Using Deep Neural Network for In-Vehicle Network Security — 2016"
[5] https://journals.nstu.ru/vestnik/download_article?id=3366 ".. , .. — — 2014"
[6] https://storage.tusur.ru/files/425/-1005__.___..pdf " .., .. — — 2013"
[7] https://www.sciencedirect.com/science/article/pii/S1877705814003579 "Halenar Igor, Juhasova Bohuslava, Juhas Martin, Nesticky Martin — Application of Neural Networks in Computer Security — 2013"
[8] http://psta.psiras.ru/read/psta2011_3_3-15.pdf " .., .., .., .. — — 2011"
[9] https://pdfs.semanticscholar.org/94f8/e1914ca526f53e9932890a0356394f9806f8.pdf "E.Kesavulu Reddy — Neural Networks for Intrusion Detection and Its Applications — 2013"
[10] http://elib.bsu.by/bitstream/123456789/179040/1/119-122.pdf " .., .., .., .. — — 2004"
[11] https://www.sans.org/reading-room/whitepapers/detection/application-neural-networks-intrusion-detection-336 "SANS Institute — Application of Neural Networks to Intrusion Detection — 2001"
[12] http://dom8a.ru/cupnewyear2014/prezentation/evlanenkova_paper.pdf " . — — ?"
[13] https://www.science-education.ru/pdf/2014/6/1632.pdf " .., .., .. — — ?"
[14] https://www.researchgate.net/publication/309038723_A_review_of_KDD99_dataset_usage_in_intrusion_detection_and_machine_learning_between_2010_and_2015 "Atilla Özgür — A review of KDD99 dataset usage in intrusion detection and machine learning between 2010 and 2015 — 2016"
[15] http://citforum.ru/security/internet/ids_overview/ " .., .. — : — 2009"
[16] https://moluch.ru/conf/tech/archive/5/1115/ " .. — . . — 2011"
[17] https://papers.nips.cc/paper/893-a-growing-neural-gas-network-learns-topologies.pdf "Fritzke B. — A Growing Neural Gas Network Learns
Topologies — 1993"
[18] https://www.mql5.com/ru/articles/163 " . — , MQL5 — 2010"
[19] https://www.researchgate.net/publication/4202425_An_incremental_growing_neural_gas_learns_topologies "Prudent Y., Ennaji A. — An incremental growing neural gas learns topologies — 2005"
[20] https://cyberleninka.ru/article/v/modifitsirovannyy-algoritm-rastuschego-neyronnogo-gaza-primenitelno-k-zadache-klassifikatsii " .., .. — , — 2014
[21] http://www.booru.net/download/MasterThesisProj.pdf "Jim Holmström — Growing Neural Gas
Experiments with GNG, GNG with Utility and Supervised GNG — 2002"