How fever kills cells

When a certain temperature is exceeded, the cell becomes unusable and dies. One of the simplest explanations for such intolerance to heat is that the proteins necessary for life - those that extract energy from food or sunlight, defend themselves from intrusion, destroy waste, etc. - often have a surprisingly accurate form. Beginning as long threads, they are then twisted in the form of spirals, "hairpins for hair" and other forms dictated by the sequence of their components. And these forms play a huge role in their activities. But when the temperature begins to rise, the bonds holding the protein structures are broken: first, the weakest, and then the strong. It is logical that the pervasive loss of protein structure should be lethal, but until recently the details of exactly how this kills overheated cells were unclear.
Now, biophysics from the Zurich Swiss Technical School have studied the behavior of each protein in the cells of four different organisms as the temperature rises. This study and a rich set of collected data, published in the journal Science , showed that at a temperature sufficient for the death of a cell — human, or Escherichia coli, E. coli — only a few key proteins are destroyed. Moreover, the abundance of proteins in the cells was unexpectedly related to their stability. Research has allowed scientists to quickly get acquainted with the fundamental rules by which the work of proteins and their ordering are built, and the consequences of which, as it became clear, extend much further than mere death from heat.
Paola Picotti , a biophysicist who led the work, explained that the experiments were repelled by old and unresolved questions: why do some cells survive at high temperatures and others die? Thermus thermophilus happily lives in hot springs and in domestic heaters [at an optimum temperature of 65 ° C - approx. trans.], whereas E. coli cells wither at temperatures above 40 ° C. Convincing evidence suggests that the matter here is in the different stability of the proteins of these organisms. But keeping track of the protein found in a living cell, which would be an ideal method of learning, is very inconvenient. Isolating protein in vitro does not give all the answers, because inside the body, proteins get together and affect each other's chemistry, or they support each other in the required form. To understand what exactly and why is falling apart, it is necessary to observe the squirrels while they are still affecting each other.
How heat destroys proteins
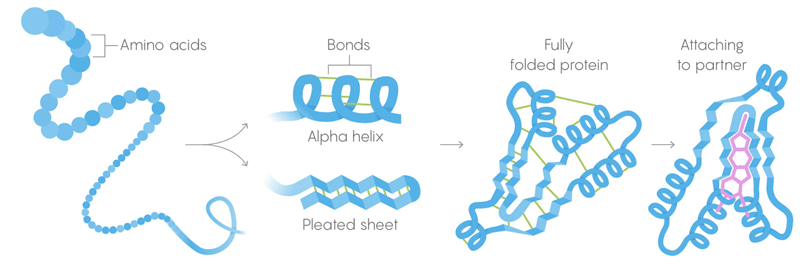
1) The primary structure of proteins is a long chain of amino acids combined into sequences defined by genes.
2) The secondary structure is an amino acid that also coagulates in a configuration held by weak intermolecular bonds.
3) Tertiary structure - weak bonds that stabilize the arrangement of straight and twisted sections of the three-dimensional structure of the protein. Their location allows the protein to connect with the desired molecules.
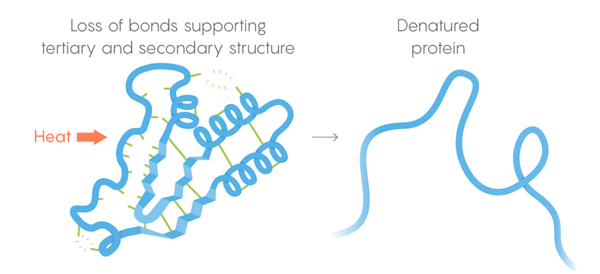
Heat death. Weak bonds lose their ability to retain tertiary and secondary structures, and the protein denatures, that is, unfolds. But not all proteins unfold at the same temperature - the environment of the protein in the cell can give it additional stabilization.
To solve the problem, the team developed an automated observation procedure. They cut the cells and heated their contents in stages, releasing the enzymes that separated the proteins at each stage. These enzymes are especially well cut unfolded proteins, so researchers on the basis of the remnants of cut proteins could judge which proteins collapsed at a given temperature. In this way, they managed to construct denaturation curves for each of the thousands of proteins studied. The arcs of the curves go from the intact structure of the protein at a comfortable temperature for it to a fully developed state at high temperature. To search for differences between the curves of different species of living creatures, experiments were carried out on human cells, E. coli, T. thermophilus and yeast. “The research was excellent,” said Alan Drummond, [Allan Drummond], a biologist at the University of Chicago, referring to both the scale and the accuracy of the process.
During the observations it was clearly seen that the proteins of all living things do not all unfold at once as the temperature rises. “We saw that only a small subset of proteins collapsed at the earliest stages,” said Picotti, “and these were key proteins.” In a diagram with interweave binding, the most fragile proteins from this small subset often have a large number of bonds, which means that they affect many of the processes that take place in cells. “Without these proteins, cells cannot work,” said Picotti. “When they disappear, the whole network collapses.” And along with it, obviously, the life of the cell also stops.
This paradox — the most important proteins are the most fragile — may be a reflection of how evolution created them to perform the corresponding tasks. If a protein has many roles, its instability and tendency to unfold and re-collapse can be an advantage, because it can allow it to take different forms suitable for different tasks. “Many of these key proteins are very flexible, which makes them less stable,” but it also gives them the ability to bind to different target molecules in the cell, explained Picotti. “Most likely, this is how they cope with their functions. This is a compromise. ”
After a closer look at E. coli, for which the collected data were the most qualitative, the researchers found a connection between the abundance of protein - the number of copies in a cell - and its stability. The more copies of a protein a cell makes, the more temperature is required to destroy it. It turns out that a large number of copies does not correlate with protein criticality for survival. Some key proteins are very rare. This connection between abundance and reliability confirms the idea put forward by Drummond ten years ago - the cellular system that makes proteins has a tendency to make mistakes from time to time. The error usually destabilizes the protein. If this protein is found to be widespread, and such a protein appears in a cell a hundred or a thousand times a day, then improperly folded copies produced in large quantities can clog the cell. Thus, it would be beneficial for the body to evolve so that the most common proteins would be the most stable, which is confirmed by the data obtained by the Picotti team.
To understand the qualities of the protein that make it stable, the researchers compared the E. coli and T. Thermophilus data. The E. coli proteins began to fall apart at 40 ° C, and almost completely degraded at 70 ° C. But at this temperature, the T. thermophilus proteins were just beginning to experience discomfort — some of them kept their shape at 90 ° C. The team found that T. thermophilus proteins were usually shorter, and some types of protein forms and components were more common in the most stable ones.
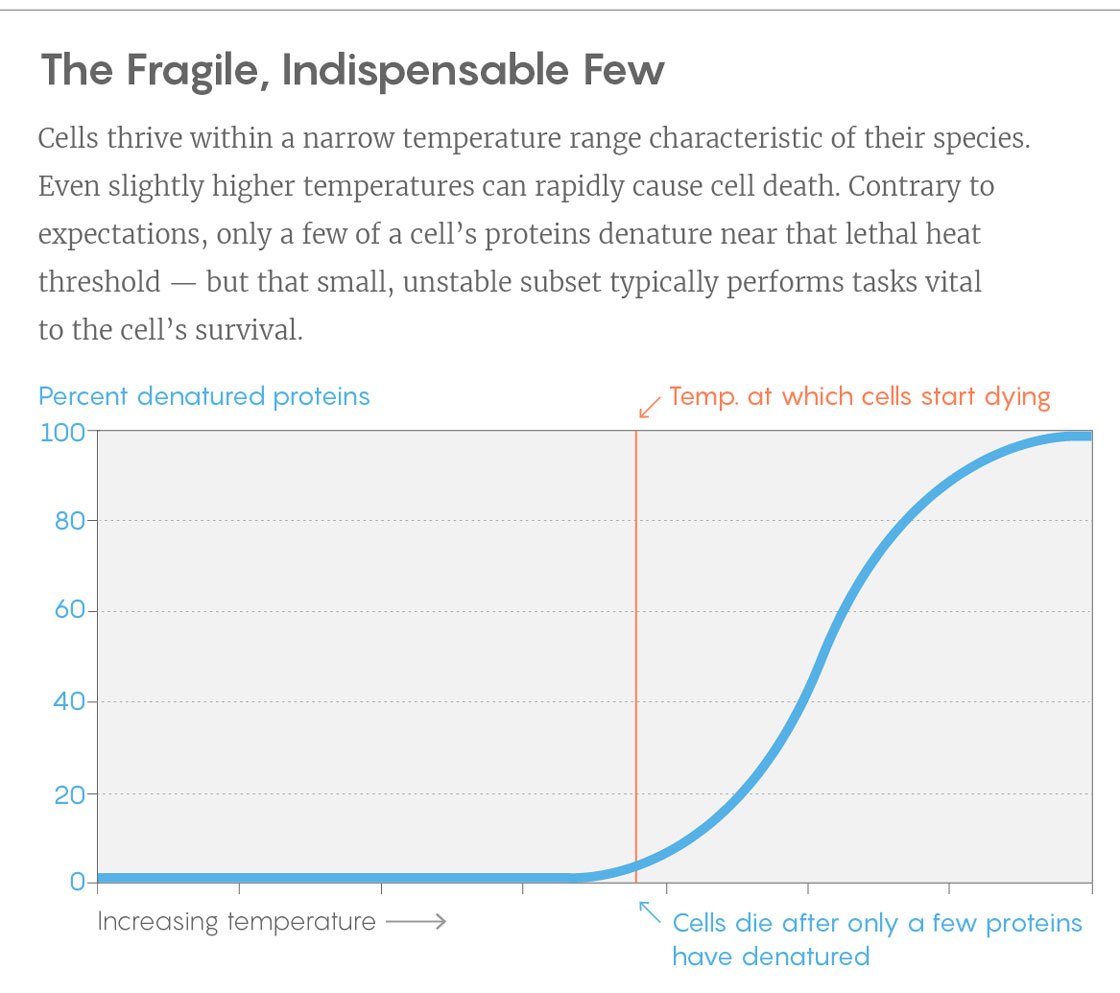
An example of a curve from the experiment. Vertically - the percentage of unfolded proteins, horizontally - the temperature. The vertical bar is the temperature at which the cells begin to die. For this you need to turn around just a few key squirrels.
Discoveries can help researchers develop proteins whose stability is tailored to their objectives. In many industrial processes where bacteria are used, a rise in temperature increases and returns - but soon enough, the bacteria begin to die from the heat. It would be interesting to find out if we can stabilize the bacteria by making key proteins more resistant to temperature, said Picotti.
The abundance of information about how easily certain proteins unfold, greatly pleased some biologists. The probability of its aggregation directly depends on the stability of a protein: the appearance of lumps of unextended proteins sticking to each other. Protein aggregates can turn into a cell nightmare and interfere with key tasks. For example, they are accused of having some serious neurological problems, such as Alzheimer's disease, in which plaques of unfolded proteins clog the brain.

Paola Picotti
But this does not mean that aggregation occurs only in organisms suffering from certain diseases. On the contrary, the researchers realized that it is possible, it happens all the time, and that healthy cells have methods with which they can cope with it. “I think that this phenomenon is increasingly recognized as very common,” said Michele Vendruscolo, a biochemist at the University of Cambridge. “Most proteins do not fold properly and aggregate inside cells. The most important thing that the Picotti team set is the length of time in which any selected protein is in an expanded state. This time determines the degree of possible protein aggregation. ” Some proteins almost never turn around and do not aggregate, others behave this way under certain conditions, while others do it all the time. According to the biochemist, a detailed description of proteins in the new work will greatly facilitate the study and understanding of these differences between proteins. Some of the denaturation curves indicate that their proteins aggregate after they are unfolded. “They managed to track both stages, both deployment and subsequent aggregation,” said Vendruscolo. “That's the beauty of this research.”
And although many scientists are interested in aggregates because of the damage they cause, some look at this phenomenon from a different point of view. Drummond says that it is becoming clear that some aggregates are not just pieces of garbage dangling in a cage. They contain active proteins that continue to perform their functions.
Imagine that you can see smoke rising from a building from afar, says Drummond. Around the building you see certain figures, and you imagine that these are bodies taken from the ruins. But if you get closer, you may find that they are living people who have escaped from a burning building, waiting for the incident to end. So it turns out with the study of aggregates, says Drummond: the researchers find that the proteins in the aggregates are not victims, but survivors. “Now a new field of science is emerging, growing at an explosive pace,” he says.
The clumping of proteins may not be a sign of damage, but a way for the protein to maintain its function in a difficult situation. It can, for example, protect them from the environment. And when conditions improve, proteins can leave the aggregates and roll up again. “Their shape changes with temperature in such a way that, at first glance, this seems like a wrong folding,” says Drummond. “But this has some other meaning.” In an article in the journal Cell from 2015, he and his colleagues identified 177 yeast proteins, which retained their functions after they hit the aggregates. In a paper published in March, this team described that if you change one of the proteins so that it cannot aggregate, this leads to serious problems in the functioning of the cell.
Overall, the paper claims that proteins are surprisingly dynamic structures. At first they may seem like hard machines working on fixed tasks for which one particular form is suitable. But in fact, proteins can take several different forms during their normal work. And at the right time, their form may change so much that it may seem as if they are deteriorating, although in fact they are on the contrary strengthened. At the molecular level, life can be a permanent connection and separation of bonds.
All Articles